Fig. 3.
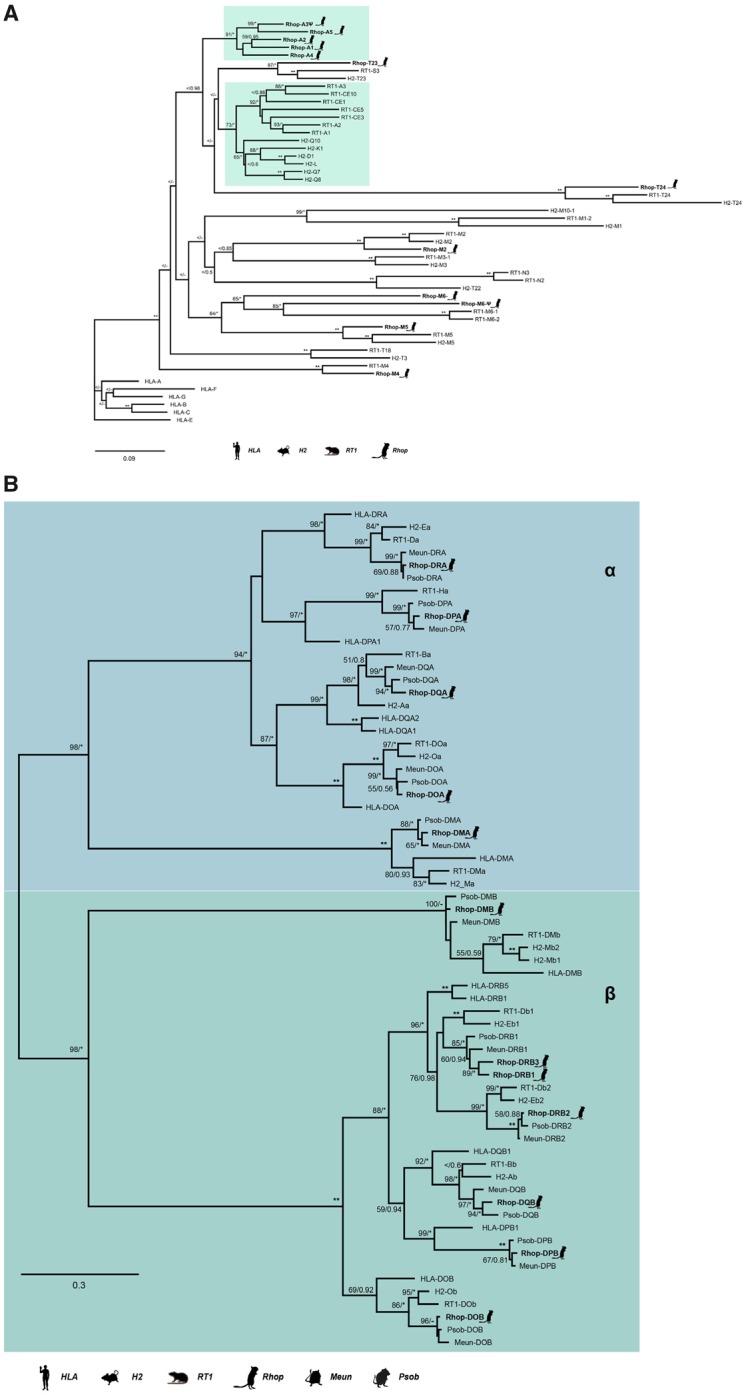
—ML phylogenies of MHCI and MHCII genes. (A) A maximum likelihood (ML) phylogeny of nucleotide sequences containing the three α domains of MHCI was created using RAxML with 100× topology and 500× bootstrap replicates. A MrBayes phylogeny with 20,000,000 generations and 25% burn-in was also created and the posterior probabilities added to the RAxML phylogeny. About 12 of the 16 great gerbil sequences were used in the analysis and are marked with a gerbil silhouette and in bold lettering. The remaining four MHCI sequences were excluded from the phylogenetic analyses due to missing data exceeding the set threshold of 50%. The clusters containing MHCIa (classical MHCI) and the closest related MHCIb genes are marked by teal boxes. Putative pseudogenes are denoted with ψ. (B) A ML phylogeny of nucleotide sequences containing the α and β domains of MHCII α and β genes was created using RAxML with 100× topology and 500× bootstrap replicates. A MrBayes phylogeny with 20,000,000 generations and 25% burn-in was also created and the posterior probabilities added to the RAxML phylogeny. Great gerbil genes are indicated with bold lettering and by silhouettes. The 12 great gerbil MHCII genes located in the genome assembly cluster accordingly with the orthologs of human, mouse, rat, sand rat, and Mongolian gerbil. The Rhop-DRB duplication (Rhop-DRB3) cluster closely with the Rhop-DRB1 and other DRB1 orthologs with good support. The nomenclature of MHCII genes in Gerbillinae is in concordance with the recommendations of the MHC Nomenclature report (Ballingall et al. 2018). BS/PP; *, BS 100 or PP > 0.96; **, BS of 100 and PP>0.97; <, support values below 50/0.8; -, node not present in Bayesian analysis.
