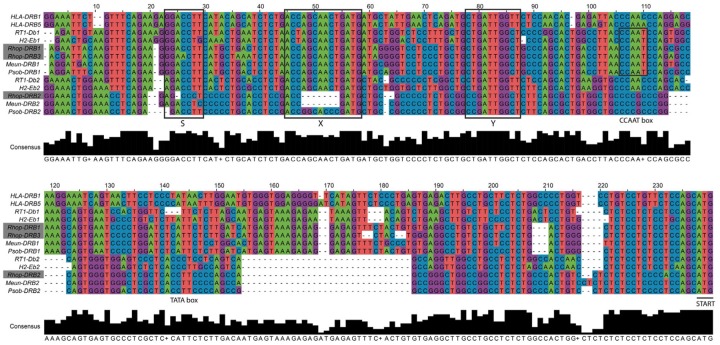
Fig. 4.
—Alignment of the DR locus β1 and β2 proximal promoters. Sequences of the proximal promoters of β1 and β2 genes of the DR locus (E locus in mouse and D locus in rat) were aligned in MEGA7 (Kumar et al. 2016) using MUSCLE with default parameters. The resulting alignment was edited manually for obvious misalignments and transferred and displayed in Jalview (Waterhouse et al. 2009). For visualization purposes only, the alignment was further edited in Adobe Illustrator (CS6), changing colors of the bases and adding boxes to point out the S–X–Y motifs. The three copies of DRB genes located in the great gerbil genome are marked with gray boxes. The alignment shows clear similarities of the proximal promoter region of Rhop-DRB1 and Rhop-DRB3 to the other rodent and human β1 promoter sequences. For the DRB2 genes, two deletions are shared among all rodents in the alignment as well as additional indels observed in Gerbillinae members. Most notably, both great gerbil and Mongolian gerbil have deletions of half the X box, whereas sand rat X box sequences in that same position are highly divergent from the otherwise conserved sequence seen in the alignment.