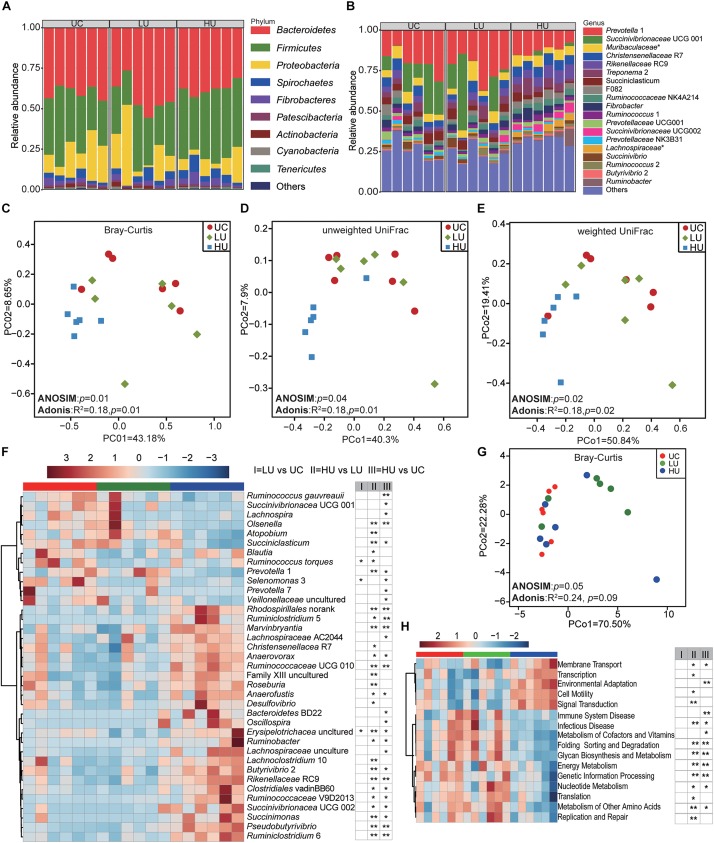
FIGURE 2.
Bacterial community compositions and potential function in the rumen solid fractions under the three treatments. Bacterial compositions in the rumen solid fractions of the UC, LU, and HU treatments at the phylum (A) and genus (B) levels. PCoA revealing the separation of the microbial communities in the rumen solid fractions among the three treatments based on Bray–Curtis dissimilarity matrix (C), unweighted UniFrac distance (D) and weighted UniFrac distance (E). Heatmap (F) showing significant differences of bacterial genera in the solid fractions among the UC, LU, and HU treatments. PCoA (G) plot revealing differences in predicted microbial functions based on Bray–Curtis dissimilarity matrix. Heatmap (H) revealing the differences of the predictive function profiles at KEGG level 2 in the rumen solid fractions among the three treatments. * p < 0.05, ** p < 0.01. UC, basal diet with no urea; LU, basal diet supplemented with a low concentration of urea (10 g/kg DM); HU, basal diet supplemented with a high concentration of urea (30 g/kg DM).