Abstract
Variants in the EYA4 gene are known to lead to autosomal dominant non-syndromic hereditary hearing loss, DFNA10. To date, 30 variants have been shown to be responsible for hearing loss in a diverse set of nationalities. To better understand the clinical characteristics and prevalence of DFNA10, we performed genetic screening for EYA4 mutations in a large cohort of Japanese hearing loss patients. We selected 1,336 autosomal dominant hearing loss patients among 7,408 unrelated Japanese hearing loss probands and performed targeted genome enrichment and massively parallel sequencing of 68 target genes for all patients. Clinical information of cases with mutations in EYA4 was gathered and analyzed from medical charts. Eleven novel EYA4 variants (three frameshift variants, three missense variants, two nonsense variants, one splicing variant, and two single-copy number losses) and two previously reported variants were found in 12 probands (0.90%) among the 1,336 autosomal dominant hearing loss families. The audiometric configuration of truncating variants tends to deteriorate for all frequencies, whereas that of non-truncating variants tends to show high-frequency hearing loss, suggesting a new correlation between genotype and phenotype in DFNA10. The rate of hearing loss progression caused by EYA4 variants was considered to be 0.63 dB/year, as found in this study and previous reports.
Subject terms: Genetic testing, Genetics research
Introduction
Hearing loss is the most common sensory disorder and more than 50% of cases of congenital or early onset hearing loss are caused by genetic factors1. Regarding hereditary hearing loss, over 100 genes are known to be causative based on genetic analysis [Hereditary Hearing Loss Homepage: https://hereditaryhearingloss.org/ accessed at August 2019], and this identification has been accelerated by the recent progress in genome sequencing technology. Hereditary hearing loss patterns vary between autosomal dominant, autosomal recessive, X-linked, and mitochondrial. Autosomal dominant non-syndromic hearing loss (ADNSHL) is often seen in postlingual hearing loss patients, and is the cause for approximately 20–25% of all hereditary hearing loss2. Currently, 67 causative genes for ADNSHL have been identified3.
The EYA4 gene (OMIM* 603550), located on chromosome 6q22.3-q23.2, for DFNA10 (OMIM# 601316) was first identified in American and Belgian hearing loss patients in 20014. EYA4 encodes eye absent 4 protein, a member of the EYA family of proteins. It is a transcriptional activator and is considered to be required for proper eye development as well as for the maturation and maintenance of the organ of Corti5. It has been reported that Eya4 is expressed in early stage otic vesicles, largely confined to the upper cochlear duct, in rodents. These cells later form the stria vascularis, Reissner’s membrane, spiral limbus, and organ of Corti. Eya4 is also expressed in the spiral ganglion neurons and organ of Corti in marmosets; however, the expression pattern in the human cochlea remains unknown6. The EYA4 protein is comprised of 639 amino acids with 2 functional domains. The C-terminal domain, which is composed of 271 residues and is named eyaHR (alternatively called the eya domain or eya homology domain 1), is highly conserved among EYA family proteins, and a more divergent proline-serine-threonine (PST)-rich transactivation domain is located at the N-terminus (eyaVR).
To date, 30 variants in the EYA4 gene and a partial or whole deletion of the EYA4 allele have been reported as a cause of ADNSHL in various ethnic groups, including the American, Belgian, Dutch, Korean, Chinese, Swedish, German, Australian, Hungarian, Philippine, Italian, and Japanese populations4,5,7–31. These previous studies have described the clinical phenotypes of patients with EYA4 variants; however, the detailed characteristics of the hearing loss, such as its progressiveness or severity, remain unclear. In addition, the genotype-phenotype correlation is also yet to be clarified.
In this study, we sought to elucidate the variant spectrum of the EYA4 gene and prevalence of EYA4-associated hearing loss in the Japanese population, and to obtain a more precise description of the clinical features of EYA4-associated hearing loss.
Materials and Methods
Study subjects
A total of 7,408 probands from unrelated Japanese hearing loss families were enrolled from 67 otolaryngology departments across Japan participating in the present study between February 2012 and October 2017. The hereditary patterns of the hearing loss in the probands’ families were autosomal dominant in 1,336, autosomal recessive/sporadic in 4,898, and unknown inheritance pattern in 1,174 cases. Written informed consent was obtained from all patients (or from their next of kin, caretaker, or legal guardian in case of minors or children). This study was approved by the Shinshu University Ethical Committee as well as the respective Ethical Committees of the other participating institutions listed below. Akita University Ethical Committee, Iwate Medical University Ethical Committee, Tohoku Rosai Hospital Ethical Committee, Fukushima Medical University Ethical Committee, Yamagata University Ethical Committee, Dokkyo Medical University Ethical Committee, TAKASAKI Ear Nose & Throat Clinic Ethical Committee, Niigata University Ethical Committee, Tokyo Medical University Ethical Committee, Jikei University Ethical Committee, Toranomon Hospital Ethical Committee, Kitasato University Ethical Committee, International University of Health and Welfare Mita Hospital Ethical Committee, National Rehabilitation Center for Persons with Disabilities Ethical Committee, Keio University Ethical Committee, Hamamatsu University Ethical Committee, Shiga University Ethical Committee, Shiga Medical Center for Children Ethical Committee, Osaka University Ethical Committee, Kobe City Medical Center General Hospital Ethical Committee, Hyogo College of Medicine Ethical Committee, Kyoto Prefectural University Ethical Committee, Okayama University Ethical Committee, Yamaguchi University Ethical Committee, Ehime University Ethical Committee, Kyushu University Ethical Committee, Kanda ENT Clinic Ethical Committee, Nagasaki University Ethical Committee, Miyazaki University Ethical Committee, Kagoshima University Ethical Committee, Ryukyus University Ethical Committee, Sapporo Medical University Ethical Committee, Tohoku University Ethical Committee, Jichi Medical University Ethical Committee, Gunma University Ethical Committee, Jyuntendo University Ethical Committee, Yokohama City University Ethical Committee, Mejiro University Ethical Committee, Saitama Medical University Ethical Committee, Abe ENT clinic Ethical Committee, Tokyo Medical Center Institute of Sensory Organs Ethical Committee, Jichi University Saitama Medical Center Ethical Committee, Aichi Children’s Health Medical Center Ethical Committee, Chubu Rosai Hospital Ethical Committee, Kyoto University Ethical Committee, Mie University Ethical Committee, Kansai Medical University Ethical Committee, Kobe University Ethical Committee, Osaka Medical Center and Research Institute for Maternal and Children Health Ethical Committee, Wakayama Medical University Ethical Committee, Kouchi University Ethical Committee, Hiroshima University Ethical Committee, Hiroshima City Hiroshima Citizen Hospital Ethical Committee, Fukuoka University Ethical Committee, Kurume University Ethical Committee, National Defense Medical College Ethical Committee, Tokai University Ethical Committee, Hokkaido University Ethical Committee, Kanagawa Children’s Medical Center Ethical Committee, Tokyo Medical and Dental University Ethical Committee, Hirosaki University Ethical Committee, Tokyo Metropolitan Children’s Medical Center Ethical Committee, Hakodate central general hospital Ethical Committee, Osaka Red Cross Hospital Ethical Committee, Hiroshima Prefectural Hospital Ethical Committee, Nara Medical University Ethical Committee, and Tsukuba University Ethical Committee. All methods were performed in accordance with the Guidelines for Genetic Tests and Diagnoses in Medical Practice of the Japanese Association of Medical Sciences and the Declaration of Helsinki as required by Shinshu University.
Clinical evaluations
The onset age of hearing loss and the degree of progressiveness were analyzed based on the medical charts of the probands and their family members harboring the same EYA4 variants. Pure-tone average (PTA) was calculated from the audiometric thresholds at four frequencies (0.5, 1, 2, and 4 kHz). The severity of hearing loss was divided into mild (PTA: 20–40 dB HL), moderate (41–70 dB HL), severe (71–95 dB HL), and profound (>95 dB HL). Asymmetric hearing loss was defined as a difference in PTA of over 10 dB between the right and left ears. The audiometric configurations were categorized into low-frequency, mid-frequency (U-shaped), high-frequency, flat type, and deaf as reported previously32.
Amplicon resequencing and variant annotation
Amplicon libraries were prepared using an Ion AmpliSeq™ Custom Panel for 68 genes reported to cause non-syndromic hereditary hearing loss (ThermoFisher Scientific, MA, USA), in accordance with the manufacturer’s instructions. The detailed protocol has been described elsewhere33. MPS was performed with an Ion Torrent Personal Genome Machine (PGM) system using an Ion PGM™ 200 Sequencing Kit and an Ion 318™ Chip (ThermoFisher Scientific). The sequence data were mapped against the human genome sequence (build GRCh37/hg19) with a Torrent Mapping Alignment Program. After sequence mapping, the DNA variant regions were piled up with Torrent Variant Caller plug-in software. After variant detection, their effects were analyzed using ANNOVAR software34,35. The missense, nonsense, insertion/deletion and splicing variants were selected from among the identified variants. Variants were further selected as less than 1% of: (1) the 1,000 genome database, (2) 6,500 exome variants, (3) the Human Genetic Variation Database (a dataset for 1,208 Japanese exome variants), and (4) 333 in-house Japanese normal hearing controls. This filtering process was performed using our original database software described elsewhere36. The pathogenicity of selected variants was evaluated by ACMG (American College of Medical Genetics) standards and guidelines37. For missense variants in particular, functional prediction software, including Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping (PolyPhen2), LRT, Mutation Taster, Mutation Assessor, Functional Analysis through Hidden Markov Models (FATHMM), RadialSVM, LR, and CADD, were used through the ANNOVAR software program34,35. Direct sequencing was utilized to confirm the selected variants.
Copy number analysis in the MPS database
We employed our recently published specialized copy number variation (CNV) detection method for Ion AmpliSeqTM sequencing that utilizes multiplex PCR-based targeted genome enrichment38. The depth of coverage information for each amplicon was used for copy number analysis. After normalization, the relative read depths of amplicons were visualized as described previously38.
Variant prioritization
EYA4 was reported as a genetic cause for autosomal dominant inherited hearing loss, thus, we selected hearing loss patients from apparently autosomal dominant families. Among 1,336 autosomal dominant hearing loss families, we further selected the families with candidate EYA4 variants. The criteria for the selection process were (1) the EYA4 variant was classified into “pathogenic”, “likely pathogenic” or “uncertain significance” and (2) there were no candidate variants in the other 67 genes reported to cause hearing loss. Based on the ACMG guidelines, we regarded “pathogenic” and “likely pathogenic” variants as strong candidates for EYA4-associated hearing loss. In addition, we listed the “variants of uncertain significance” identified during the filtering procedure described above in Table 1. However, we removed “variants of uncertain significance” with a CADD Phred score of less than 20, or identified in some control databases as being of “unlikely causative”. The CADD Phred score threshold used in this study was <20 as all of the previously reported EYA4 pathogenic variants were predicted to have a CADD Phred score of 23.5 or more (e.g., the lowest CADD Phred score for c.978C > G is 23.5), so we employed 20 as threshold to allow a safety margin. In addition, we also removed the c.1790delT and c.1886_1899del variants as unlikely causative variants because nonsense-mediated mRNA decay was not presumed to be triggered from the location of the variants. Finally, we selected 12 variants as causative and performed a more detailed hearing loss phenotype analysis.
Table 1.
All EYA4 variants found in this study.
| # | RefSeq ID | Nucleotide Change | Amino Acid Change | Exon | Domain | Genomic position (GRCh37.p5) | SIFT | PolyPhen2_HVIR | PolyPhen2_HVAR | LRT | Mut _Taster | Mut _Assessor | FATHMM | Meta SVM | Meta LR | CADD_Phred | Allele Frequency (Exac03) | ACMG criteria (supporting evidence) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Likely causative | |||||||||||||||||||
| 1 | NM_004100 | c.222_223del | p.T74fs | 5 | V | 133769262 | 0 | Likely Pathogenic (PVS1 + PM2) | |||||||||||
| 2 | NM_004100 | c.498delG | p.Q166fs | 8 | V | 133783533 | 0 | Likely Pathogenic (PVS1 + PM2) | |||||||||||
| 3 | NM_004100 | c.517C > T | p.Q173X | 8 | V | 133783552 | 0.843 | 0.810 | 39.000 | 0 | Likely Pathogenic (PVS1 + PM2 + PP3) | ||||||||
| 4 | NM_004100 | c.580 + 1G > A | intron 8 | V | 133783616 | 0.810 | 26.200 | 0 | Likely pathogenic(PVS1 + PM2 + PP3) | ||||||||||
| 5 | NM_004100 | c.910delC | p.P304fs | 11 | V | 133789809 | 0 | Likely Pathogenic (PVS1 + PM2) | |||||||||||
| 6 | NM_004100 | c.988C > T | p.Q330X | 12 | V | 133802618 | 0.843 | 0.810 | 47.000 | 0 | Likely Pathogenic (PVS1 + PM2 + PP3) | ||||||||
| 7 | NM_004100 | c.1109G > C | p.R370P | 13 | E | 133804171 | 0.912 | 0.899 | 0.971 | 0.629 | 0.810 | 0.888 | 0.975 | 0.995 | 0.984 | 34.000 | 0 | VUS (PM2 + PP3) | |
| 8 | NM_004100 | c.1177C > T | p.Q393X | 13 | E | 133804239 | 0.843 | 0.810 | 46.000 | 0.000008253 | 0.00009639(AFR) | Pathogenic (PVS1 + PS1 + PP3) | |||||||
| 9 | NM_004100 | c.1216G > C | p.G406R | 14 | E | 133827268 | 0.912 | 0.764 | 0.693 | 0.629 | 0.810 | 0.907 | 0.808 | 0.897 | 0.885 | 29.800 | 0 | VUS (PM2 + PP3) | |
| 10 | NM_004100 | c.1663G > C | p.A555P | 18 | E | 133844240 | 0.784 | 0.899 | 0.916 | 0.843 | 0.810 | 0.865 | 0.975 | 0.989 | 0.985 | 32.000 | 0 | VUS (PM2 + PP3) | |
| 11 | NM_004100 | CNV | 133782193–133789881 | Likely pathogenic (PVS1 + PM2) | |||||||||||||||
| 12 | NM_004100 | CNV | 133756417–133852199 | Likely pathogenic (PVS1 + PM2) | |||||||||||||||
| Unlikely causative | |||||||||||||||||||
| 13 | NM_004100 | c.278T > C | p.M93T | 6 | V | 133777694 | 0.153 | 0.090 | 0.127 | 0.305 | 0.810 | 0.065 | 0.623 | 0.514 | 0.500 | 14.76 * | 0 | VUS (PM2) | |
| 14 | NM_004100 | c.887C > T | p.S296L | 11 | V | 133789786 | 0.721 | 0.548 | 0.533 | 0.629 | 0.537 | 0.741 | 0.853 | 0.816 | 0.812 | 23.600 | 0.00001651 ** | 0.0002(EAS) ** | VUS (PP3) |
| 15 | NM_004100 | c.936G > T | p.L312F | 11 | V | 133789835 | 0.491 | 0.764 | 0.764 | 0.843 | 0.810 | 0.805 | 0.862 | 0.800 | 0.842 | 23.000 | 0.00001651 ** | 0.0002(EAS) ** | VUS (PP3) |
| 16 | NM_004100 | c.995C > T | p.P332L | 12 | V | 133802625 | 0.348 | 0.764 | 0.739 | 0.843 | 0.810 | 0.684 | 0.953 | 0.970 | 0.972 | 27.900 | 0.000008264 ** | 0.0001(EAS) ** | VUS (PS1 + PP3) |
| 17 | NM_004100 | c.1790delT | p.V597fs | 19 | E | 133846343 | 0 | Pathogenic (PVS1 *** + PS1 + PM2) | |||||||||||
| 18 | NM_004100 | c.1886_1899del | p.A629fs | 20 | E | 133849909 | 0 | VUS (PVS1 *** + PM2) | |||||||||||
Abbreviation: V, variable region; E, Eya domain.
*CADD score is low.
**MAF is too high.
***These variants are unlikely causative because nonsense-mediated mRNA decay is not presumed to be triggered from the location of the variants (see Discussion section).
Results
Identified variants and the frequency of EYA4-associated hearing loss
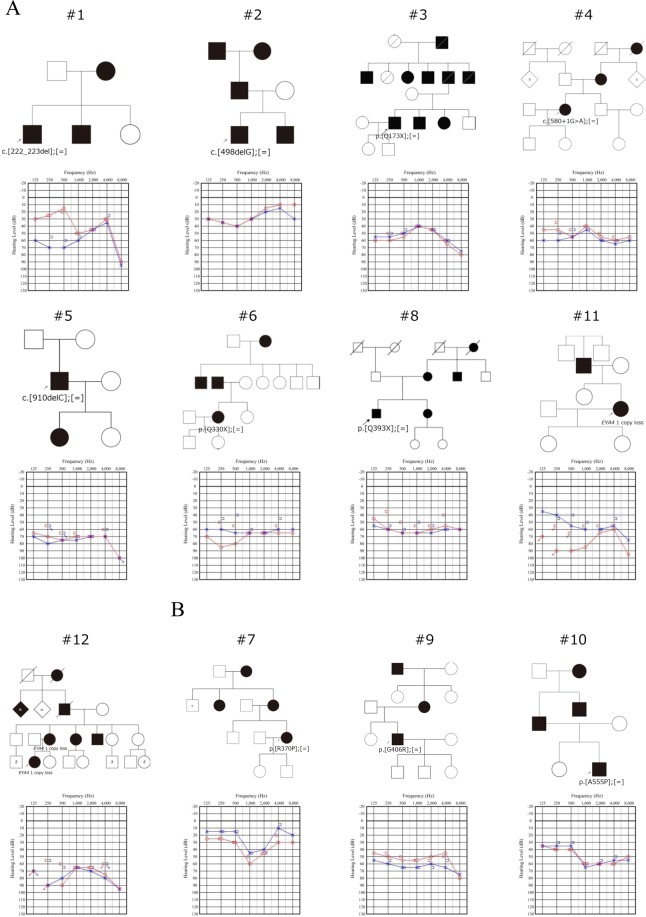
Among the 1,336 probands with ADNSHL, we identified 12 (0.90%) who carried a possible EYA4 pathogenic variant (Table 1, Fig. 1). These 12 probands did not show any pathogenic variants or candidate variants in the 67 previously reported deafness genes apart from EYA4. Among the 12 candidate variants, eleven were novel, and one was previously reported. Three of them were missense variants, three were frameshift insertion/deletion variants, three were nonsense variants, one was a splicing variant, and two were copy number losses. Six of them were located in the eyaVR (amino acids 0–369) and four were located in eyaHR (amino acids 370–639). The one previously reported variant was classified as “Pathogenic”. Eight variants were classified as “Likely pathogenic” according to the ACMG guidelines, whereas three remained as “variants of uncertain significance (VUS)”. Further, none of the 12 variants was found in the Japanese 333 in-house controls (666 control alleles), and none of the three VUS variants was observed in the ExAC03 database.
Figure 1.
Pedigrees and audiograms of twelve families who carried a possible EYA4 pathogenic variant identified in this study. Filled symbols indicate affected individuals. Arrows indicate probands in each family. Unfortunately, we could not obtain DNA samples from family members except #12, so we could not perform family segregation analysis.
In addition to the above 12 causative EYA4 variants, we also identified 6 variants in the EYA4 gene from our cohort (Table 1), but we regarded these 6 variants as unlikely to be causative. Three missense variants, c.887C > T, c.936G > T, and c.995C > T were identified in the ExAC03 database over 0.0001, suggesting these three variants were not causative variants. In addition, the c.278T > C variant was predicted to be an unlikely causative variant from its low CADD score (CADD Phred score 14.76). Furthermore, the c.1790delT and c.1886_1899del variants were regarded as unlikely causative variants as nonsense-mediated mRNA decay was not presumed to be triggered from the location of the variants (these variants were located in the final exon or one exon before the final exon). From these results, the pathogenicity of these six variants was unclear. Thus, we performed further detailed clinical characteristic analysis for 12 patients with causative EYA4 variants.
Clinical characteristics of the EYA4-associated hearing loss patients identified in this study
Table 2 summarizes the clinical characteristics of the 12 individuals with EYA4 variants. The onset age of hearing loss varied markedly from 5 to 61 years old; however, the onset of hearing loss in most cases was in the second decade or later. Mild to moderate hearing loss was observed in many cases, but two cases showed severe hearing loss. Asymmetric hearing loss was observed in two individuals (Family 1, 11). Audiometric configurations in these patients included low-frequency type in 6 ears, mid-frequency type in 2 ears, high-frequency type in 4 ears, and flat type in 12 ears. All individuals had noticed a progression in their hearing loss.
Table 2.
Clinical features of affected family members with EYA4 mutations found in this study.
| Family No. | Nucleotide Change | Amino Acid Change | Sex | Domain | HL | Pure-tone audiometry | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Onset (y) | Progression | Tested age (y) | PTA (R/L) | Severity (R/L) | Audiometric configuration (R/L) | |||||
| Truncating variant | ||||||||||
| 1 | c.222_223del | p.T74fs | M | V | 61 | Yes | 64 | 35/52.5 | mild/moderate | HF/LF |
| 2 | c.498delG | p.Q166fs | M | V | 13 | Yes | 13 | 23.8/26.3 | mild/mild | LF/LF |
| 3 | c.517C > T | p.Q173X | M | V | 48 | Yes | 53 | 51.3/48.8 | moderate/moderate | flat/flat |
| 4 | c.580 + 1G > A | F | V | 45 | Yes | 47 | 52.5/56.3 | moderate/moderate | flat/flat | |
| 5 | c.910delC | p.P304fs | M | V | 30 | Yes | 61 | 71.3/72.5 | severe/severe | flat/flat |
| 6 | c.988C > T | p.Q330X | F | V | 16 | Yes | 34 | 68.8/63.8 | moderate/moderate | flat/flat |
| 8 | c.1177C > T | p.Q393X | M | E | 26 | Yes | 36 | 61.3/63.8 | moderate/moderate | flat/flat |
| 11 | CNV | F | 25 | Yes | 51 | 75/57.5 | severe/moderate | LF/HF | ||
| 12 | CNV | F | 13 | Yes | 47 | 73.8/76.3 | severe/severe | LF/LF | ||
| Non-truncating variant | ||||||||||
| 7 | c.1109G > C | p.R370P | F | E | 30 | Yes | 41 | 41.3/27.5 | moderate/mild | MF/MF |
| 9 | c.1216G > C | p.G406R | M | E | 5 | Yes | 77 | 51.3/63.8 | moderate/moderate | flat/flat |
| 10 | c.1663G > C | p.A555P | M | E | 25 | Yes | 37 | 55/53.8 | moderate/moderate | HF/HF |
Abbreviations: HL, hearing loss; HF, high-frequency hearing loss; MF, mid-frequency hearing loss; LF, low-frequency hearing loss; N/A, not available.
Analysis of hearing deterioration in the EYA4-associated hearing loss patients
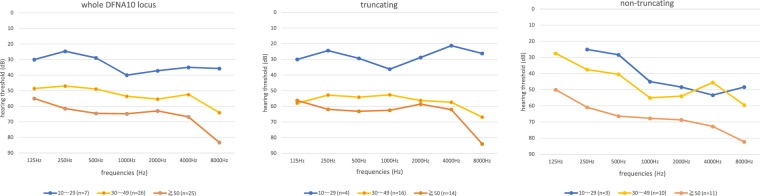
To elucidate more precisely the type of hearing loss and rate of hearing deterioration, we collected the hearing thresholds of our patients. In addition, we also collected the hearing thresholds described in previous reports5,16,20,27. From our study results, we incorporated the hearing thresholds for 12 cases in this analysis (six cases considered to be unlikely causative variants were excluded from this analysis). We compared the hearing thresholds of patients with truncating variants to those with non-truncating variants (missense variants) including the patients in this study and previously reported cases (Fig. 2). As a result, the patients with truncating variants revealed a flat-type hearing loss that deteriorated in all frequencies, whereas the patients with non-truncating variants showed high-frequency hearing loss. We also analyzed the rate of hearing deterioration by using the patients in this study and previously reported case results (Fig. 3) and found that the average rate of progression in PTA was 0.63 dB/year (95%CI: 0.41–0.85 dB/year).
Figure 2.
Audiometric configuration of EYA4-associated hearing loss. The left panel indicates the audiometric configuration of all of EYA4-associated HL. The center panel indicates the audiometric configuration of EYA4-associated HL with truncating variants. The right panel indicates the audiometric configuration of the EYA4-associated HL patients with non-truncating variants. This analysis was performed using the hearing thresholds from the 12 patients identified in this study and previously reported cases (n = 33). The blue line indicates 10–29 y. o. patients, the yellow line indicates 30–49 y. o. patients and the orange line indicates patients 50 y. o. or above.
Figure 3.
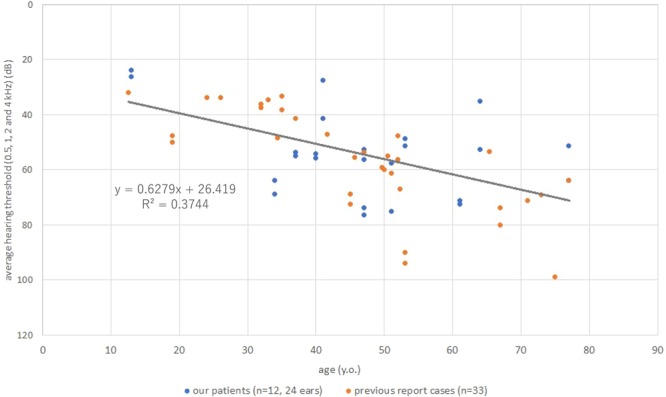
The estimated progression rate in PTA of EYA4-associated HL patients in this study and previous reports. The average progression rate in PTA was 0.63 dB/year.
Discussion
In this report, we analyzed 1,334 ADNSHL patients and identified 12 candidate variants for EYA4-associated hearing loss. This is the largest population studied for EYA4-associated hearing loss to date. The prevalence of EYA4-associated hearing loss in ADNSHL was 0.90% (12/1,334 cases) in the Japanese population. This prevalence is slightly less than those of other ADNSHL genes such as KCNQ4, TECTA, POU4F3, and WFS1. KCNQ4 is one of the most frequently observed responsible genes for ADNSHL in the Japanese population, and its prevalence is 6.6%39. Likewise, the prevalence of ADNSHL caused by TECTA variants is 2.9%, 2.7% for POU4F3 variants, and 2.5% for WFS1 variants40–42.
The responsible genes for ADNSHL differ among ethnic groups. For example, KCNQ4 is the most frequent causative gene for ADNSHL in the Japanese population, whereas TECTA is the most frequent causative gene in the American population43. One plausible reason of this difference among populations is the effect of founder or recurrent mutations. Indeed, the variant of KCNQ4; c.211delC, which is commonly observed in Japanese ADNSHL patients, was reported to be caused by a founder effect39. Most of the EYA4 variants found in this report were novel (we summarized the clinical features and identified variants of previous reports in Table 3) and the identified variants differed among patients. Only one variant (p.Q393X) was identified in a Korean patient9. According to this result, most of the EYA4 variants are not recurrent. From these results, it appears difficult to find EYA4 variants among autosomal dominant hereditary hearing loss patients by various genotyping analysis methods such as Invader assay or microarray, thus MPS is useful for identifying rare causative variants such as those in the EYA4 gene in ADNSHL patients.
Table 3.
Summary of the clinical features associated with EYA4 mutations from previous studies.
| Mutation type | Nucleotide Change | Exon/Intron | Amino Acid Change | Domain | Onset | Progression | Severity of HL | Audiometric Configuration | Family Origin | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Missense or nonsense | c.152C > T | exon 4 | p.S51F | Variable region | N/A | N/A | N/A | N/A | America | Sloan-Heggen, 20167 |
| c.511G > C | exon 8 | p.G171R | Variable region | 6–50 | Yes | mild to severe | flat, HF | China | Liu, 20155 | |
| c.863C > A | exon 11 | p.S288X | Variable region | N/A N/A | N/A N/A | N/A moderate | N/A flat | Korea | Beak, 20128 Kim, 20159 | |
| c.866C > T | exon 11 | p.T289M | Variable region | N/A | N/A | N/A | N/A | America | Miszalski-Jamka, 201710 | |
| c.978C >G | exon 12 | p.F326L | Variable region | N/A | N/A | N/A | N/A | Korea | Choi, 201311 | |
| c.1109G > A | exon 13 | p.R370H | Eya domain | N/A | N/A | N/A | N/A | Philippines | Truong, 201912 | |
| c.1111G > A | exon 13 | p.V371M | Eya domain | N/A | N/A | N/A | N/A | Belgium | Sommen, 201613 | |
| c.1154C > T | exon 13 | p.S385L | Eya domain | N/A | N/A | N/A | N/A | Italy | Cesca, 201814 | |
| c.1177C > T | exon 13 | p.Q393X | Eya domain | N/A | N/A | moderate | HF | Korea | Kim, 20159 | |
| c.1223G > A | exon 14 | p.R408H | Eya domain | N/A | N/A | N/A | N/A | America | Miszalski-Jamka, 201714 | |
| c.1301T > A | exon 15 | p.I434K | Eya domain | N/A | N/A | N/A | N/A | China | Tan, 201415 | |
| c.1643C > G | exon 18 | p.T548R | Eya domain | 19–37 | Yes | mild to profound | flat, HF | China | Sun, 201516 | |
| c.1759C > T | exon 19 | p.R587X | Eya domain | N/A | N/A | N/A | N/A | Belgium | Wayne, 20014 | |
| c.1810G > T | exon 19 | p.G604C | Eya domain | N/A | N/A | N/A | N/A | Netherlands | Neveling, 201317 | |
| c.1834A > T | exon 19 | p.K612X | Eya domain | N/A | N/A | mild to moderate | flat, HF | China | Hu, 201818 | |
| Splicing | c.84-2A > G | intron 3 | N/A | N/A | N/A | N/A | China | Chen, 201619 | ||
| c.1282-12T > A | intron 14 | 6–30 s | Yes | moderate to severe | flat, HF | Australia | Hildebrand, 200720 | |||
| c.1341-19T > A | intron 15 | N/A | N/A | N/A | N/A | Germany | Vona, 201421 | |||
| c.1739-1G > A | intron 18 | 50 | N/A | N/A | N/A | America | Cirino, 201722 | |||
| Deletion | c.464delC | exon 8 | Variable region | N/A | N/A | N/A | N/A | Sweden | Neveling, 201317 | |
| c.1194delT | exon 14 | p.Met401Trpfs*3 | Eya domain | 20 s | Yes | moderate to severe | HF | Korea | Choi, 201623 | |
| c.1790delT | exon 19 | Eya domain | 35 | Yes | moderate | flat | Japan | Iwasa, 201624 | ||
| Insertion | c.579_580insTACC | exon 8 | p.Asp194Tyrfs*52 | Variable region | N/A | N/A | N/A | N/A | Sweden | Frykholm, 201525 |
| c.614dupA | exon 9 | Variable region | N/A | N/A | N/A | N/A | China | Huang, 201526 | ||
| c.1026_1027dupAA | exon 12 | Variable region | N/A | N/A | N/A | N/A | America | Wayne, 20014 | ||
| c.1048_1049dupAA | exon 12 | Variable region | 20s-40s | Yes | moderate to severe | flat, HF | America | Makishima, 200727 | ||
| c.1115_1118dupTTGT | exon 13 | Eya domain | N/A | N/A | N/A | N/A | Hungary | Pfister, 200228 | ||
| Gross deletion | 10.4 Mb promoter and exon 1,2 | N/A | N/A | N/A | N/A | Japan | Abe, 200929 | |||
| 4846 bp intron 10 | N/A | N/A | N/A | N/A | America | Schönberger, 200530 | ||||
| 9 Mb exon 4-20 | N/A | N/A | N/A | N/A | Poland | Dutrannoy, 200931 |
Abbreviations: HL, hearing loss; HF, high-frequency hearing loss; N/A, not available.
In previous reports, the audiometric configuration for EYA4-associated hearing loss was a gradual high-frequency hearing loss or a flat-type hearing loss44. Further, no genotype-phenotype correlation was identified in previous reports. Kim et al. reported that no genotype-phenotype correlation existed for EYA4-associated hearing loss9. In their report, they analyzed only 87 ADNSHL patients, and identified only two patients carrying EYA4 variants. In this study, we analyzed 1,334 ADNSHL patients, and identified 12 candidate EYA4 variants. We also analyzed the detailed audiometric configurations of 12 patients identified in this study and previously reported cases and identified a genotype-phenotype correlation. High-frequency hearing loss was observed in patients with non-truncating EYA4 variants, whereas flat-type hearing loss was observed in patients with truncating EYA4 variants. In contrast, there were no significant differences in the severity of hearing loss among the different types of variants and/or variant locations (domain).
We also analyzed the rate of hearing deterioration in EYA4-associated hearing loss patients identified in this study and previously reported cases. The rate of progression of hearing loss caused by EYA4 was considered to be 0.63 dB/year (95%CI: 0.41–0.85 dB/year). In previous reports on ADSNHL hearing loss, the progression rate for the POU4F3 gene was 0.5–0.9 dB/year41, that for MYO6 was 2.0 dB/year45, and that for ACTG1 was 2.0–6.0 dB/year46, and the results in this study suggests that the rate of hearing loss progression caused by EYA4 may be relatively mild.
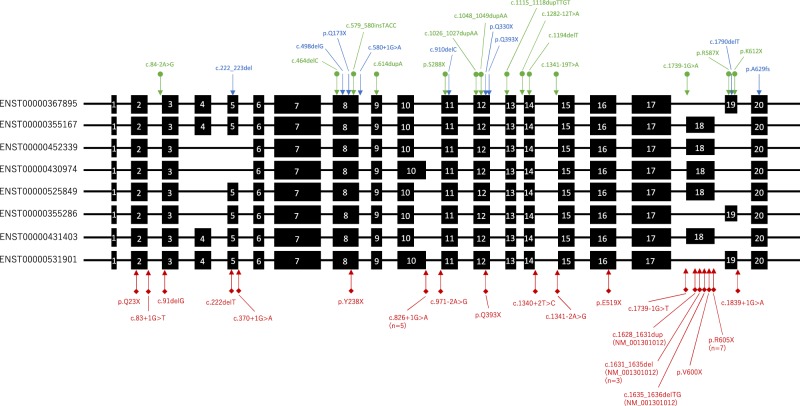
In this study, we identified nine truncating variants including two EYA4 copy number loss cases. Thus, we speculated that the mechanism of EYA4-associated hearing loss was haploinsufficiency. In the gnomAD database (https://gnomad.broadinstitute.org/gene/ENSG00000112319), a non-negligible number of truncating variants were identified in large control populations. The probability of a loss of function intolerant score (pLI score) was 0.05. This low score may mean the loss of function in this gene is tolerant and without pathogenicity. However, most of the loss of function variants were located in specific exons that only included some splicing variants and were seldom observed in other exons (Fig. 4). From these observations, we hypothesized that these specific isoforms may not be expressed in the inner ear or may not play an important role in hearing function. It is unknown which isoforms are expressed in the human inner ear. As another hypothesized mechanism, loss of function variants in the gnomAD database were accumulated in the second to last exon, and these variants might not trigger nonsense-mediated mRNA decay. Thus, these loss of function variants may not cause hearing loss. The identified truncating variants, except for c.1790delT, were located in the exons which were included in all isoforms carried. The prevalence of loss of function variants in the EYA4 gene was 19 among about 250,000 alleles in the gnomAD database, but 9 among 2,672 alleles in this study. The summarized odds ratio between our hearing loss cohort vs. gnomAD was 44.469 (95%CI: 20.495–96.490). This result also supports haploinsufficiency as the mechanism underlying EYA4-associated hearing loss. The patient who carried c.1790delT, located in specific exons (truncating variants accumulated in the exon in gnomAD) suffers from an enlarged vestibular aqueduct, and this phenotype was not matched with hearing loss caused by EYA4 mutations. For these reasons, we classified this variant (c.1790delT) as “unlikely causative”.
Figure 4.
The location of truncating variants identified in this study, previous reports and the genomAD database. The schema shows alternative splicing variants of the EYA4 gene. Black boxes indicate exons present in each transcript variant and black lines indicate introns. Blue arrows indicate truncating variants identified in this study. Green arrows indicate truncating variants identified in previous reports. Red arrows indicate truncating variants in the genomAD database.
In conclusion, we performed MPS analysis of large cohort of 1,334 ADNSHL patients and successfully identified 12 novel and promising pathogenic variants. Based on this, we estimated the incidence of EYA4-associated hearing loss was 0.90% in Japanese families with autosomal dominant hearing loss. The audiometric configuration of truncating variants tended to exhibit flat-type, whereas that of non-truncating variants tended to be high-frequency hearing loss, suggesting a novel genotype-phenotype correlation in DFNA10.
Acknowledgements
We thank the participants of the Deafness Gene Study Consortium for providing samples and clinical information [Nishio and Usami, 2015]. This study was aided by a Health and Labor Sciences Research Grant for Research on Rare and Intractable Diseases and Comprehensive Research on Disability Health and Welfare from the Ministry of Health, Labour and Welfare of Japan (S.U. H29-Nanchitou(Nan)-Ippan-031), a Grant-in-Aid from Japan Agency for Medical Research and Development (AMED) (S.U. 16kk0205010h001, 15ek0109114h001), a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (S.U. 15H02565) and a Grant-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (H.M.15K10747).
Author contributions
Study conception & design: H.M., S.N. and S.U.; Acquisition of data: J.S., K.O., K.O., S.I., S.M., C.O., Y.O., Y.A., M.T., N.S., S.A., Y.S., H.S., T.I. and N.U.; Bioinformatics analysis: H.M. and S.N.; Sanger sequencing analysis: J.S., H.M. and S.N.; Data analysis and interpretation: J.S. and H.M.; Writing of the manuscript: J.S., H.M. and S.N.; Study supervision: S.U. All authors read and approved of the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Morton CC, Nance WE. Newborn Hearing Screening — A Silent Revolution. N. Engl. J. Med. 2006;354:2151–2164. doi: 10.1056/NEJMra050700. [DOI] [PubMed] [Google Scholar]
- 2.Van Laer L, et al. Autosomal Dominant Nonsyndromic Hearing Impairment. Am. J. Med. Genet. 1999;89:167–174. doi: 10.1002/(SICI)1096-8628(19990924)89:3<167::AID-AJMG7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.Meena R, Ayub M. Genetics Of Human Hereditary Hearing Impairment. J. Ayub Med. Coll. Abbottabad. 2017;29:671–676. [PubMed] [Google Scholar]
- 4.Wayne S, et al. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 2002;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- 5.Liu F, et al. Exome sequencing identifies a mutation in EYA4 as a novel cause of autosomal dominant non-syndromic hearing loss. PLoS One. 2015;10:1–11. doi: 10.1371/journal.pone.0126602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuzaki S, Hosoya M, Okano H, Fujioka M, Ogawa K. Expression pattern of EYA4 in the common marmoset (Callithrix jacchus) cochlea. Neurosci. Lett. 2018;662:185–188. doi: 10.1016/j.neulet.2017.10.030. [DOI] [PubMed] [Google Scholar]
- 7.Sloan-Heggen CM, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee K-Y, et al. Targeted massive parallel sequencing: the effective detection of novel causative mutations associated with hearing loss in small families. Orphanet J. Rare Dis. 2012;7:60. doi: 10.1186/1750-1172-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YR, et al. Evaluation of the contribution of the EYA4 and GRHL2 genes in korean patients with autosomal dominant non-syndromic hearing loss and GRHL2 genes in korean patients with autosomal dominant non-syndromic hearing loss. PLoS One. 2015;10:1–10. doi: 10.1371/journal.pone.0119443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miszalski-Jamka, K. et al. Novel Genetic Triggers and Genotype-Phenotype Correlations in Patients with Left Ventricular Noncompaction. Circ. Cardiovasc. Genet. 10 (2017). [DOI] [PMC free article] [PubMed]
- 11.Choi BY, et al. Diagnostic Application of Targeted Resequencing for Familial Nonsyndromic Hearing Loss. PLoS One. 2013;8:1–8. doi: 10.1371/journal.pone.0068692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Truong BT, et al. Exome sequencing reveals novel variants and unique allelic spectrum for hearing impairment in Filipino cochlear implantees. Clin. Genet. 2019;95:634–636. doi: 10.1111/cge.13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sommen M, et al. DNA Diagnostics of Hereditary Hearing Loss: A Targeted Resequencing Approach Combined with a Mutation Classification System. Hum. Mutat. 2016;37:812–819. doi: 10.1002/humu.22999. [DOI] [PubMed] [Google Scholar]
- 14.Cesca F, et al. A novel mutation of the EYA4 gene associated with post-lingual hearing loss in a proband is co-segregating with a novel PAX3 mutation in two congenitally deaf family members. Int. J. Pediatr. Otorhinolaryngol. 2018;104:88–93. doi: 10.1016/j.ijporl.2017.10.042. [DOI] [PubMed] [Google Scholar]
- 15.Tan M, et al. Identification of I411K, a novel missense EYA4 mutation causing autosomal dominant non-syndromic hearing loss. Int. J. Mol. Med. 2014;34:1467–1472. doi: 10.3892/ijmm.2014.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, et al. A novel mutation of EYA4 in a large Chinese family with autosomal dominant middle-frequency sensorineural hearing loss by targeted exome sequencing. J. Hum. Genet. 2015;60:299–304. doi: 10.1038/jhg.2015.19. [DOI] [PubMed] [Google Scholar]
- 17.Neveling K, et al. A Post-Hoc Comparison of the Utility of Sanger Sequencing and Exome Sequencing for the Diagnosis of Heterogeneous Diseases. Hum. Mutat. 2013;34:1721–1726. doi: 10.1002/humu.22450. [DOI] [PubMed] [Google Scholar]
- 18.Hu, S. et al. Genetic Etiology Study of Ten Chinese Families with Nonsyndromic Hearing Loss. Neural Plast. 2018 (2018). [DOI] [PMC free article] [PubMed]
- 19.Chen S, et al. Targeted Next-Generation Sequencing Successfully Detects Causative Genes in Chinese Patients with Hereditary Hearing Loss. Genet. Test. Mol. Biomarkers. 2016;20:660–665. doi: 10.1089/gtmb.2016.0051. [DOI] [PubMed] [Google Scholar]
- 20.Hildebrand MS, et al. A Novel Splice Site Mutation in EYA4 Causes DFNA10 Hearing Loss. Am. J. Med. Genet. 2007;Part A:1599–1604. doi: 10.1002/ajmg.a.31860. [DOI] [PubMed] [Google Scholar]
- 21.Vona B, et al. Targeted next-generation sequencing of deafness genes in hearing-impaired individuals uncovers informative mutations. Genet. Med. 2014;16:945–953. doi: 10.1038/gim.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirino, A. L. et al. A Comparison of Whole Genome Sequencing to Multigene Panel Testing in Hypertrophic Cardiomyopathy Patients. Circ. Cardiovasc. Genet. 10 (2017). [DOI] [PMC free article] [PubMed]
- 23.Choi HS, Kim AR, Kim SH, Choi BY. Identification of a novel truncation mutation of EYA4 in moderate degree hearing loss by targeted exome sequencing. Eur. Arch. Oto-Rhino-Laryngology. 2016;273:1123–1129. doi: 10.1007/s00405-015-3661-2. [DOI] [PubMed] [Google Scholar]
- 24.Iwasa YI, Nishio SY, Usami SI. Comprehensive genetic analysis of Japanese autosomal dominant sensorineural hearing loss patients. PLoS One. 2016;11:1–11. doi: 10.1371/journal.pone.0166781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frykholm C, et al. Phenotypic variability in a seven-generation Swedish family segregating autosomal dominant hearing impairment due to a novel EYA4 frameshift mutation. Gene. 2015;563:10–16. doi: 10.1016/j.gene.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 26.Huang A, Yuan Y, Liu Y, Zhu Q, Dai P. A novel EYA4 mutation causing hearing loss in a Chinese DFNA family and genotype-phenotype review of EYA4 in deafness. J. Transl. Med. 2015;13:1–7. doi: 10.1186/s12967-015-0483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makishima Tomoko, Madeo Anne C., Brewer Carmen C., Zalewski Christopher K., Butman John A., Sachdev Vandana, Arai Andrew E., Holbrook Brenda M., Rosing Douglas R., Griffith Andrew J. Nonsyndromic hearing loss DFNA10 and a novel mutation ofEYA4: Evidence for correlation of normal cardiac phenotype with truncating mutations of the Eya domain. American Journal of Medical Genetics Part A. 2007;143A(14):1592–1598. doi: 10.1002/ajmg.a.31793. [DOI] [PubMed] [Google Scholar]
- 28.Pfister M, et al. A 4bp-insertion in the eya-homologous region (eyaHR) of EYA4 causes hearing impairment in a Hungarian family linked to DFNA10. Mol. Med. 2002;8:607–611. doi: 10.1007/BF03402171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe Y, et al. EYA4, deleted in a case with middle interhemispheric variant of holoprosencephaly, interacts with SIX3 both physically and functionally. Hum. Mutat. 2009;30:E946–E955. doi: 10.1002/humu.21094. [DOI] [PubMed] [Google Scholar]
- 30.Schönberger J, et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- 31.Dutrannoy V, et al. De novo 9 Mb deletion of 6q23.2q24.1 disrupting the gene EYA4 in a patient with sensorineural hearing loss, cardiac malformation, and mental retardation. Eur. J. Med. Genet. 2009;52:450–453. doi: 10.1016/j.ejmg.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Mazzoli M, et al. Recommendations for the Description of Genetic and Audiological Data for Families with Nonsyndromic Hereditary Hearing Impairment. Audiol. Med. 2003;1:148–150. doi: 10.1080/16513860301713. [DOI] [Google Scholar]
- 33.Miyagawa M, Nishio Sya, Ikeda T, Fukushima K, Usami Sichi. Massively Parallel DNA Sequencing Successfully Identifies New Causative Mutations in Deafness Genes in Patients with Cochlear Implantation and EAS. PLoS One. 2013;8:1–8. doi: 10.1371/journal.pone.0075793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J. Hum. Genet. 2008;6:2166–2171. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:1–7. doi: 10.1093/nar/gkp829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishio SY, Usami SI. The Clinical Next-Generation Sequencing Database: A Tool for the Unified Management of Clinical Information and Genetic Variants to Accelerate Variant Pathogenicity Classification. Hum. Mutat. 2017;38:252–259. doi: 10.1002/humu.23160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishio Sya, Moteki H, Usami Sichi. Simple and efficient germline copy number variant visualization method for the Ion AmpliSeqTM custom panel. Mol. Genet. Genomic Med. 2018;6:678–686. doi: 10.1002/mgg3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naito T, et al. Comprehensive Genetic Screening of KCNQ4 in a Large Autosomal Dominant Nonsyndromic Hearing Loss Cohort: Genotype-Phenotype Correlations and a Founder Mutation. PLoS One. 2013;8:e63231. doi: 10.1371/journal.pone.0063231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moteki H, et al. TECTA mutations in Japanese with mid-frequency hearing loss affected by zona pellucida domain protein secretion. J. Hum. Genet. 2012;57:587–592. doi: 10.1038/jhg.2012.73. [DOI] [PubMed] [Google Scholar]
- 41.Kitano T, et al. POU4F3 mutation screening in Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis identified novel variants associated with autosomal dominant hearing loss. PLoS One. 2017;12:1–15. doi: 10.1371/journal.pone.0177636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi M, et al. WFS1 mutation screening in a large series of Japanese hearing loss patients: Massively parallel DNA sequencing-based analysis. PLoS One. 2018;13:1–19. doi: 10.1371/journal.pone.0193359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hildebrand MS, et al. DFNA8/12 Caused by TECTA Mutations is the Most Identified Subtype of Non-syndromic Autosomal Dominant Hearing Loss. Hum. Mutat. 2012;32:825–834. doi: 10.1002/humu.21512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Leenheer EM, et al. THE DFNA10 PHENOTYPE. Ann. Otol. Rhinol. Laryngol. 2001;110:861–866. doi: 10.1177/000348940111000910. [DOI] [PubMed] [Google Scholar]
- 45.Miyagawa M, Nishio SY, Kumakawa K, Usami SI. Massively Parallel DNA Sequencing Successfully Identified Seven Families with Deafness-Associated MYO6 Mutations: The Mutational Spectrum and Clinical Characteristics. Ann. Otol. Rhinol. Laryngol. 2015;124:148S–157S. doi: 10.1177/0003489415575055. [DOI] [PubMed] [Google Scholar]
- 46.Miyagawa M, et al. Mutational Spectrum and Clinical Features of Patients with ACTG1 Mutations Identified by Massively Parallel DNA Sequencing. Ann. Otol. Rhinol. Laryngol. 2015;124:84S–93S. doi: 10.1177/0003489415575057. [DOI] [PubMed] [Google Scholar]