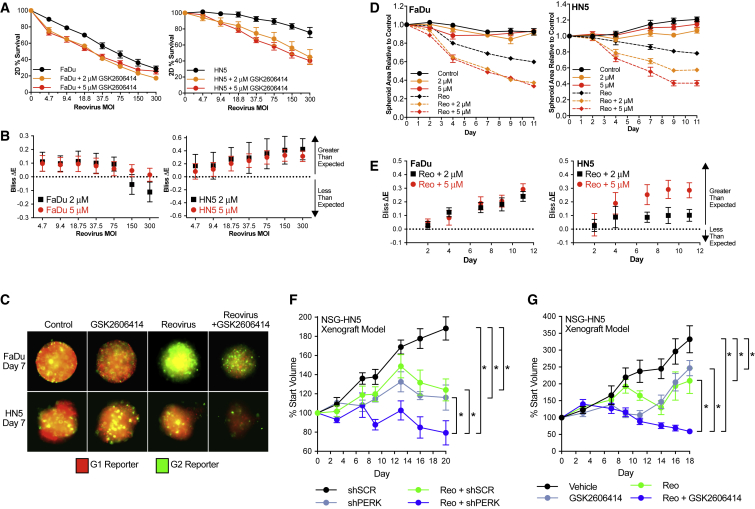
Figure 1.
GSK2606414 Sensitizes HNSCC to Reovirus
(A) Cell viability of FaDu and HN5 HNSCC cell lines was determined at 72 h by MTT assay. GSK2606414 and reovirus were added concurrently. (B) Observed cell kill versus expected cell kill for MTT assays was determined by Bliss independence analysis. Expected cell kill is calculated on the assumption that single agents are non-interacting. Positive ΔE corresponds to greater observed kill than expected, and negative ΔE less than expected. Full details are outlined in Materials and Methods. (C) FaDu and HN5 HNSCC cell lines were plated in low-attachment U-bottom plates to form 3D tumor spheroids expressing G1-mCherry and G2-AMCyan cell-cycle trackers. Spheroids were treated with GSK2606414 and reovirus concurrently. Spheroids were imaged using cell-cycle tracker fluorescence at the indicated time points. (D) Area relative to control was calculated by automated image quantification using images as shown in panel C. (E) Observed reduction versus expected reduction for 3D spheroid areas was determined by Bliss independence analysis as in (B). (F) Increased efficacy of reovirus in combination with loss of PERK was determined in vivo by Tet-inducible shRNA. HN5 cells stably expressing lentiviral Tet-pLKO-puro were injected subcutaneously into NSG mice. HN5 cells contained either scrambled shRNA (SCRsh) or PERK targeting shRNA (PERKsh). All mice received 50 mg/kg doxycycline daily by gavage with reovirus delivered by intra-tumoral injection at day zero. Tumor volumes are expressed relative to start volume. For validation of knockdown, see Figure S1. (G) Increased efficacy of reovirus in combination with GSK2606414 was determined in vivo in HN5 cells injected subcutaneously into NSG mice. Mice received 50 mg/kg GSK2606414 on days 0–4, 7–11, and 14–18 with two injections of reovirus on days 0 and 9. (A and D) Data are ±SEM of at least three independent experiments. Bliss independence analysis in (B) and (E) shown with 95% confidence intervals. In vivo statistical analysis shown between groups in (F) and (G) by unpaired t test, *p < 0.05 of area under curve comparison for individual tumors.