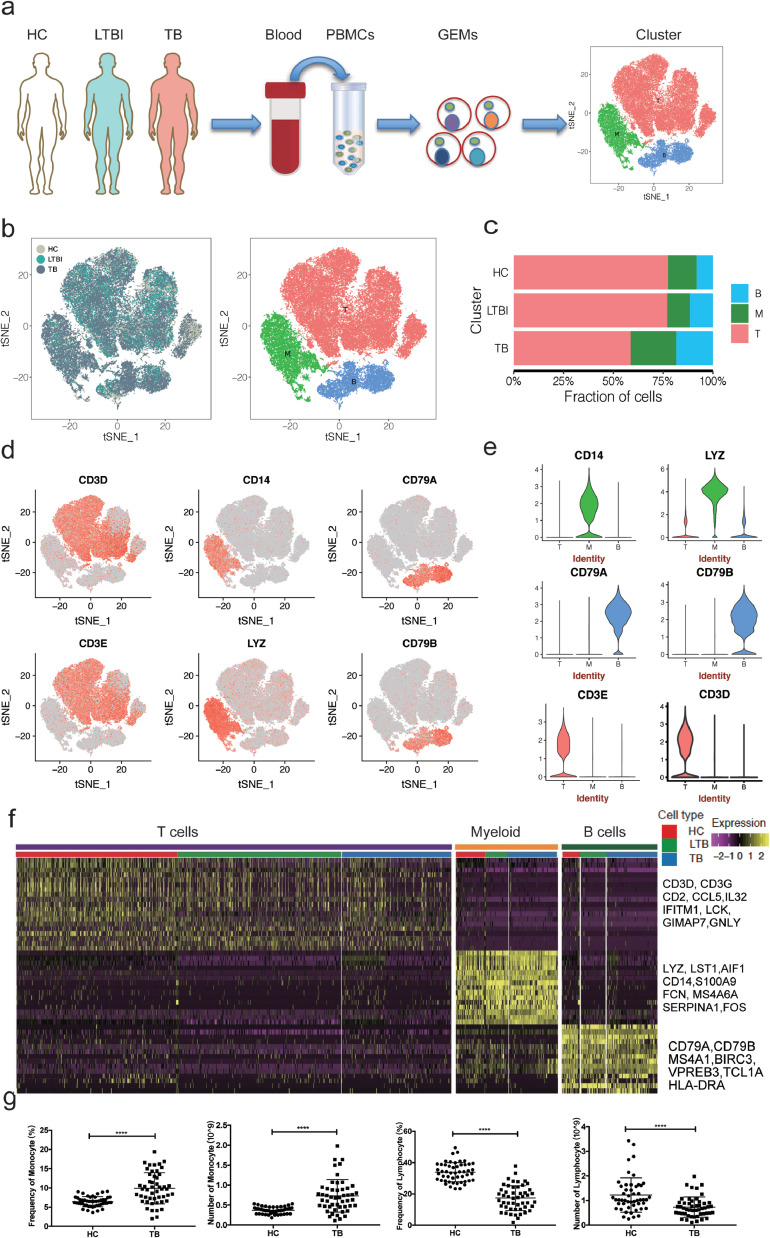
Fig. 1.
Single-cell transcriptional profiling of PBMCs from HC, LTBI and TB. (a) Schematic of experimental workflow for defining and comparing PBMC between all three donor groups. (b) tSNE of single cell profile with each cell color-coded for sample type and associated cell type. (c) The fraction of cells for three cell type in HC, LTBI and TB. (d)Expression of marker genes for the cell types defined above each panel. (e) The expression of known cell type discriminating genes in T, B and Myeloid from all donors.(f) Differential expression analysis was performed comparing cells from HC, LTBI and TB within T, B and Myeloid cells. Heatmaps are shown representing the up- and down-regulated genes in T, B and Myeloid cells. (g) Blood routine analysis of the numbers and frequencies of monocytes and lymphocyte from HC and TB. Unpaired t-test were used and the data represent the means ± SEM. ***P < 0. 001, ***P < 0. 0001.