Figure 3.
Discrimination of miRNA Isoforms (isomiRs) Using NIPSS
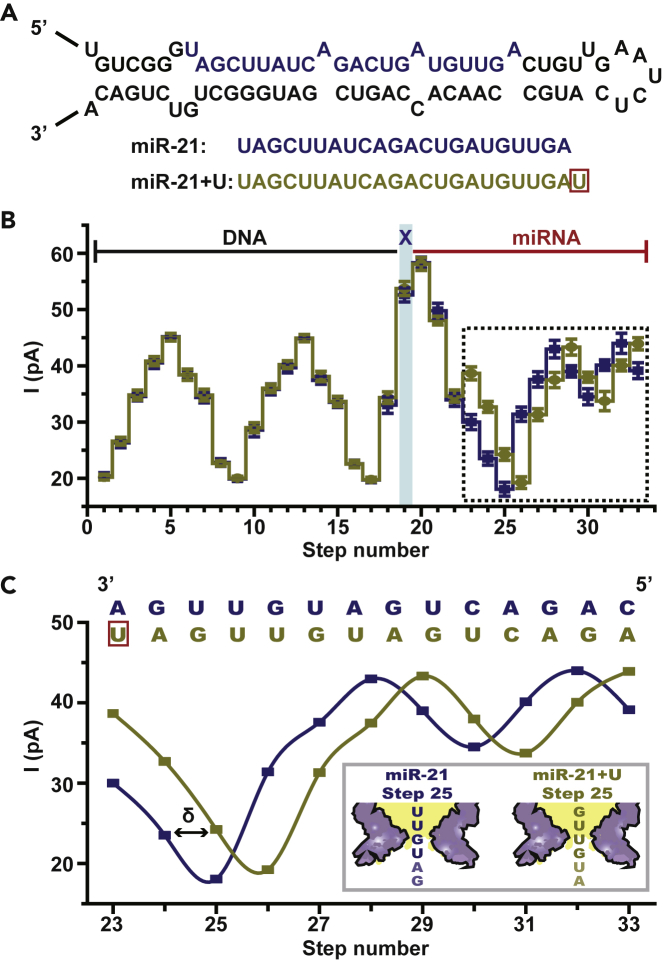
(A) miR-21 and its isoforms. The upper image demonstrates the structure of a precursor microRNA (pre-miRNA) for human miR-21. The lower image demonstrates the sequence (5′-3′) of mature miR-21 and its miR-21 + U isoform, respectively. An additional uracil (red box) exists at the 3′-end of miR-21 + U.
(B) Comparison of consensus sequencing results between DNA-miR-21 and DNA-miR-21 + U. The means and standard deviations were derived from 24 time-normalized independent events for DNA-miR-21 (blue) and DNA-miR-21 + U (brown) respectivly (Table S3). The DNA, the abasic site, and the miRNA part of the signal are marked separately. The blue strip marks the sequencing step of TCAX, which is the first sequence quadromer containing an abasic spacer encountered by NIPSS. The demonstrated statistical results show great alignment in all parts except that marked with a dashed-line box.
(C) Sequencing result shift between DNA-miR-21 and DNA-miR-21 + U. A zoomed-in view of the dashed-line box in B illustrates a shift effect of current levels caused by a nucleotide insertion of DNA-miR-21 + U in reference to DNA-miR-21. The aligned sequence context above the results demonstrates that the addition of a uracil (red box) in DNA-miR-21 + U systematically generates 1 nucleotide (marked as δ) shift. This single nucleotide result shift is also demonstrated by the schematic diagram in the image inset, which takes the results of step 25 as an example.
See also Tables S1 and S3, Figures S7 and S8.