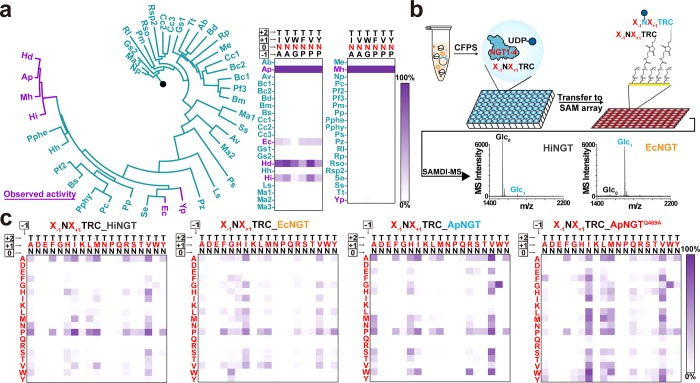
Figure 2.
GlycoSCORES screening of NGT homologues for unique peptide specificities. (a) Screening for active NGTs from putative bacterial glycosyltransferases. Forty-one putative glycosyltransferases from the CAZY database were screened against six representative peptide substrates for N-glucosyltransferase activity. The phylogenetic tree is rooted with human OGT (black dot) as the outgroup. Six enzymes showed N-glucosyltransferase activity, with strong activity from ApNGT, EcNGT, HdNGT, HiNGT, and MhNGT. Relative MS intensities of peptide substrates and glucose modified peptide products are shown when treated with 5% (v/v) CFPS NGTs and 2.5 mM UDP-Glc at 30 °C for 21 h (n = 1). (b) Scheme for GlycoSCORES workflow for discovery and characterization of the peptide specificities for the NGTs. NGTs were produced in CFPS and mixed with UDP-Glc sugar donor and cysteine-containing peptide substrates, which were then immobilized to a maleimide functionalized self-assembled monolayer and characterized by SAMDI-MS. (c) Peptide acceptor specificity comparison of HiNGT (gray), EcNGT (orange), ApNGT (blue), and ApNGTQ469A (red) using a peptide array of X–1NX+1TRC sequence. Expanded X–1NX+1(T/S)RC substrate libraries with numerical annotation are shown in Supplementary Figure 4. Enzyme concentrations were controlled to obtain a maximum yield of ∼85% in each heatmap to facilitate the comparison of specificities. The heatmap for ApNGT is taken from a previous report.36 Each heatmap shows percentage glucose modification from n = 1 experiment using the conditions: 0.42 μM CFPS HiNGT or 0.75 μM CFPS EcNGT, 30 °C for 21 h; 0.055 μM CFPS ApNGT or 0.014 μM CFPS ApNGTQ469A, 30 °C for 1 h.