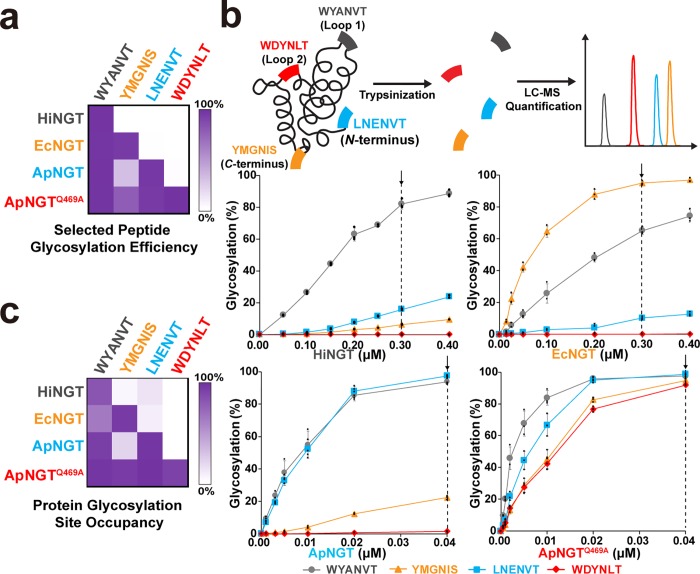
Figure 4.
Optimized GlycTag sequences showing conditional orthogonality as peptides and within a single Im7 target protein. (a) The conditional orthogonality of optimized 6-mer GlycTags. HiNGT, EcNGT, ApNGT, and ApNGTQ469A modification of selected GlycTags from Figure 3c under optimized conditions were resynthesized and reanalyzed by GlycoSCORES with purified NGTs. The modification efficiency of each peptide was calculated using individually measured RIFs (Supplementary Table 4). Experimental conditions (n = 3 individual IVGs): 0.2 μM HiNGT or 0.67 μM EcNGT, 30 °C for 21 h; 0.45 μM ApNGT or 0.1 μM ApNGTQ469A, 30 °C for 3 h. (b) Optimized 6-mer GlycTags were inserted into the N-terminus, C-terminus, and two exposed loops of the glycosylation model protein Im7, with flanking sequences of RATT-GlycTag-RAGG to facilitate trypsinization and quantitative LC-qTOF analysis. 10 μM of purified 4gIm7 bearing all four optimized GlycTags was reacted with 2.5 mM UDP-Glc and various concentrations of each purified NGT for 4 h in n = 3 IVG reactions. After modification, 4gIm7 was purified from the reaction, treated with trypsin, and analyzed by LC-qTOF. The modification efficiency (occupancy) of each site was calculated using individually measured RIFs (shown in Supplementary Table 5). A vertical dotted line in each graph denotes optimal conditions for each NGT which provide the greatest conditional orthogonality. Representative LC chromatograms and related MS spectra are presented in Supplementary Figure 13. (c) The conditional orthogonality of each optimized 6-mer GlycTag within 4gIm7 under optimized conditions. Bar graphs with errors for heatmaps in (a) and (c) are shown in Supplementary Figure 10. Similar modification patterns were observed for peptides and GlycTags within engineered 4gIm7.