Figure 1.
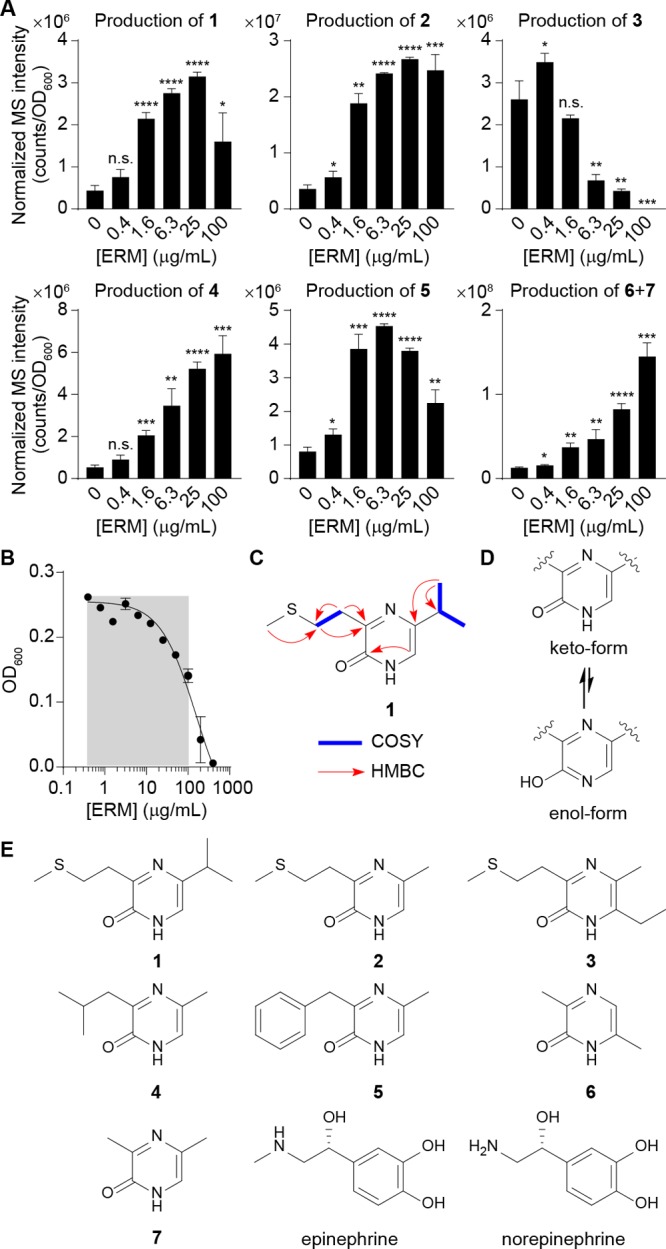
Characterization of pyrazinone metabolites differentially regulated by ribosomal inhibitory stress in E. coli. (A) Dose-dependent production of 1–7 in E. coli Nissle 1917 by erythromycin, as established by LC-MS. Compounds 6 and 7 are indistinguishable by the standard LC-MS method. ERM, erythromycin. (B) Half-maximal inhibitory concentration (IC50) analysis of E. coli Nissle 1917 in response to erythromycin. Gray area represents sublethal antibiotic range examined for metabolite production. (C) Key NMR correlations for representative pyrazinone 1. NMR spectra and analysis for all metabolites can be found in the Supporting Information. COSY, blue bold. HMBC, red arrows. (D) Keto-form is favored versus enol-form. (E) Structures of characterized bacterial metabolites (1–7) and human-derived epinephrine and norepinephrine. Revised pyrazinone tautomeric structure of DPO (7) is shown. n = 3 biological replicates. Data are mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-tailed t-test. ns, not significant.
