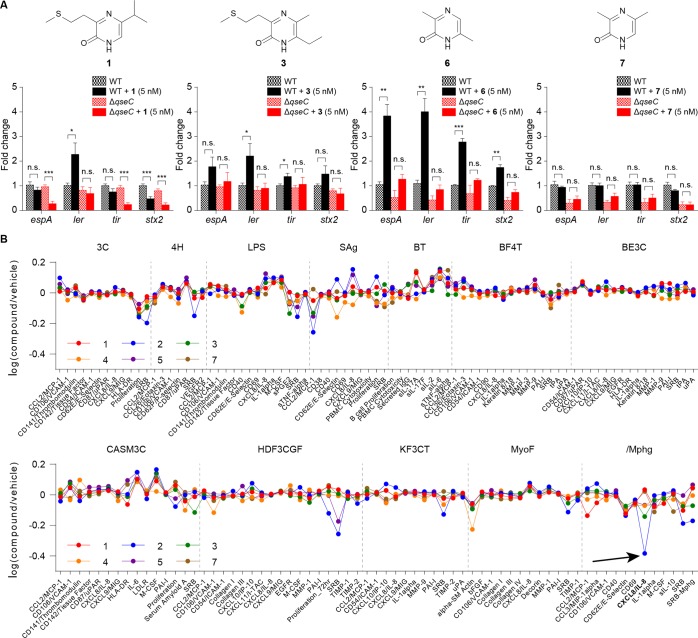
Figure 5.
Evaluation of the biological activities of metabolites 1–7. (A) qRT-PCR analysis for the expression of virulence genes from WT EHEC and ΔqseC EHEC with pyrazinones 1, 3, 6, or 7 (5 nM). n = 6 (three biological replicates and two technical replicates). Fold change was calculated relative to rpoA as an internal control. Data are mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant. Two-tailed t-test. See Figure S11 for marginal 2, 4, and 5 responses. (B) BioMAP Phenotypic Profiling assay with pyrazinones 1–5 and 7 (36 μM). Cell types and stimuli used in each system are as follows: 3C system [HUVEC + (IL-1β, TNFα, and IFNγ)], 4H system [HUVEC + (IL-4 and histamine)], LPS system [PBMC and HUVEC + LPS (TLR4 ligand)], SAg system [PBMC and HUVEC + TCR ligands (1×)], BT system [CD19+ B cells and PBMC + (α-IgM and TCR ligands (0.001×))], BF4T system [bronchial epithelial cells and HDFn + (TNFα and IL-4)], BE3C system [bronchial epithelial cells + (IL-1β, TNFα, and IFNγ)], CASM3C system [coronary artery smooth muscle cells + (IL-1β, TNFα, and IFNγ)], HDF3CGF system [HDFn + (IL-1β, TNFα, IFNγ, EGF, bFGF, and PDGF-BB)], KF3CT system [keratinocytes and HDFn + (IL-1β, TNFα, and IFNγ)], MyoF system [differentiated lung myofibroblasts + (TNFα and TGFβ)] and /Mphg system [HUVEC and M1 macrophages + zymosan (TLR2 ligand)].