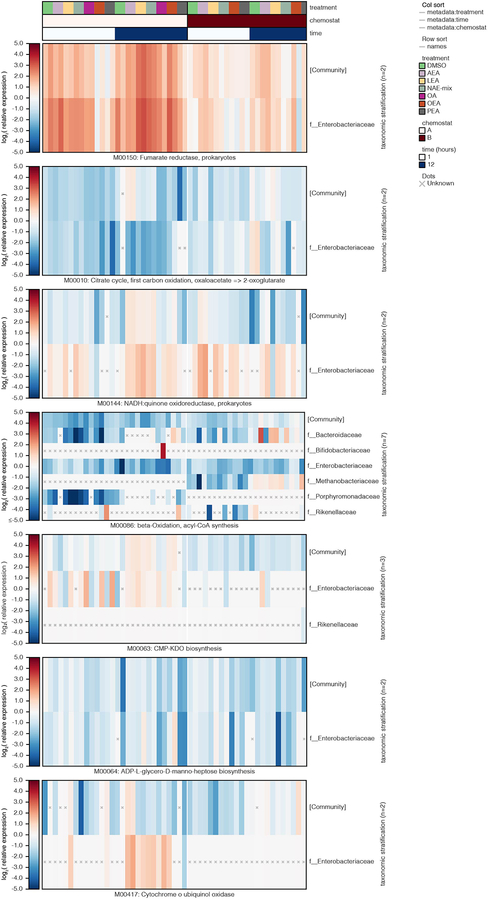
Extended Data Fig. 9. Changes in transcriptional activity (KEGG modules) in complex microbial communities in response to NAE treatment.
Log2 relative expression is shown for bacterial families contributing to a selection of KEGG modules in samples from chemostats A and B treated with DMSO control, individual NAEs, a combination of all four NAEs (denoted as NAE-mix), or oleic acid (OA). Family-level relative expression values were computed at the species level, averaged over replicates, and then averaged within-family while weighting by species abundance. Unknown (“x”) values represent cases where a function’s DNA and/or RNA abundance were zero for a given stratification (resulting in non-finite log2 relative expression). Col_sort refers to the measure (treatment, time or chemostat used to order the metadata columns). Abbreviations: PEA, palmitoylethanolamide; OEA, oleoyl ethanolamide; LEA, linoleoyl ethanolamide; AEA, arachidonoyl ethanolamide.