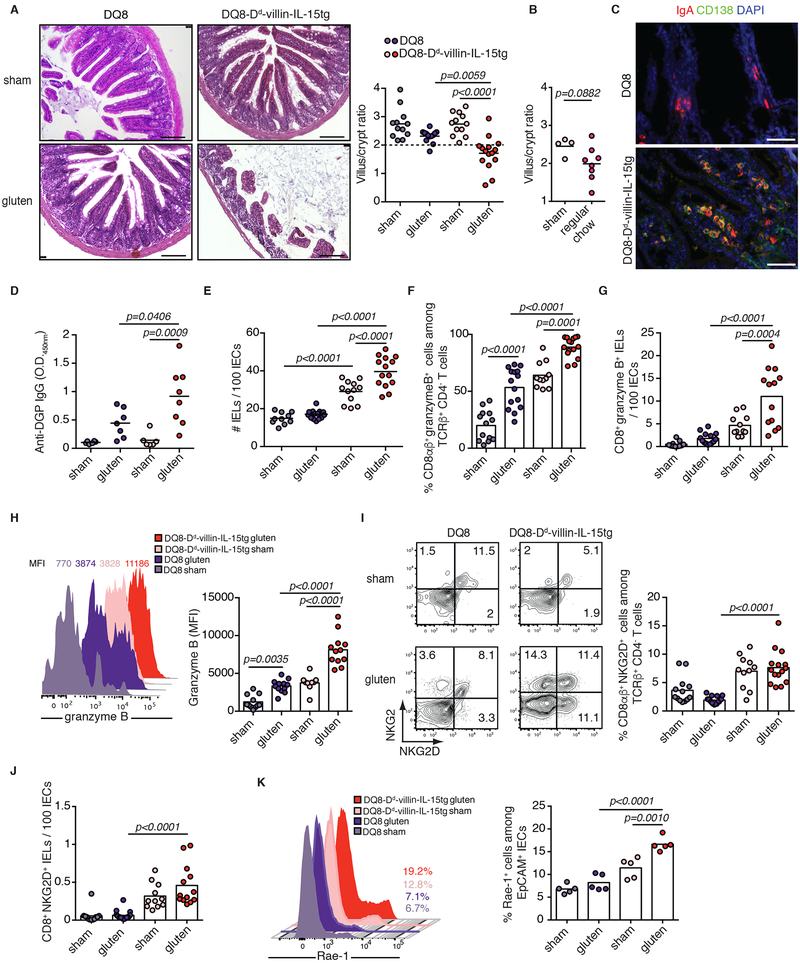
Extended Data Figure 3. Overexpression of IL-15 in HLA-humanized DQ8 mice confers susceptibility to development of coeliac disease-like features in a gluten-dependent manner.
(A-K) DQ8 and DQ8-Dd-villin-IL-15tg mice were maintained on a GFD (sham) or fed with gluten for 30 days (gluten).
(A) H&E staining of paraffin-embedded ileum sections. Scale bar, 100 μm. The graph depicts the ratio of the morphometric assessment of villous height to crypt depth. Four independent experiments shown (DQ8 sham, n=12, gluten n=13; DQ8-Dd-villin-IL-15tg sham, n=12, gluten n=15); mean; ANOVA / Tukey’s multiple comparison.
(B) Villous to crypt ratio from sham-fed DQ8-Dd-villin-IL-15tg mice and DQ8-Dd-villin-IL-15tg mice fed a standard rodent chow without supplemental gluten. Two independent experiments (sham, n=4, gluten n=8); mean; Unpaired, two-tailed, t-test.
(C) IgA (red) and CD138+ plasma cells (green) were distinguished by immunohistochemistry (IHC) staining of frozen ileum sections.
(D) Serum anti-deamidated gliadin peptides (DGP) IgG levels as measured by ELISA. Sera were collected thirty days after gluten feeding. Two independent experiments (DQ8 sham, n=6, gluten n=7; DQ8-Dd-villin-IL-15tg sham, n=6, gluten n=7); mean; ANOVA / Tukey’s multiple comparison.
(E) Quantification of IELs among IECs performed on H&E stained ileum sections. Four independent experiments (DQ8 sham, n=10, gluten n=16; DQ8-Dd-villin-IL-15tg sham, n=12, gluten n=14); mean; ANOVA / Tukey’s multiple comparison.
(F-J) The intestinal epithelium was isolated and analyzed by flow cytometry. IELs were identified as TCRβ+ CD4− CD8+ and TCRβ+ CD4− CD8αβ+ cells.
(F) Granzyme B+ IELs are indicated by percentage. Four independent experiments (DQ8 sham, n=13, gluten n=15; DQ8-Dd-villin-IL-15tg sham, n=11, gluten n=15); mean; ANOVA / Tukey’s multiple comparison.
(G) Numbers of granzyme B+ CD8+ IELs / 100 IECs. Four independent experiments (DQ8 sham, n=11, gluten n=14; DQ8-Dd-villin-IL-15tg sham, n=11, gluten n=13); mean; ANOVA / Tukey’s multiple comparison.
(H) Granzyme B MFI. Data are representative of three independent experiments shown as mean (DQ8 sham, n=11, gluten n=12; DQ8-Dd-villin-IL-15tg sham, n=9, gluten n=12). ANOVA / Tukey’s multiple comparison.
(I) NKG2D+ NKG2− IELs are indicated by percentage. Four independent experiments (DQ8 sham, n=13, gluten n=17; DQ8-Dd-villin-IL-15tg sham, n=11, gluten n=15); mean; ANOVA / Tukey’s multiple comparison.
(J) Numbers of NKG2D+ NKG2− IELs / 100 IECs. Four independent experiments (DQ8 sham, n=12, gluten n=16; DQ8-Dd-villin-IL-15tg sham, n=11, gluten n=13); mean; ANOVA / Tukey’s multiple comparison.
(K) The intestinal epithelium was isolated and analyzed by flow cytometry. Intestinal epithelial cells (IECs) were identified as EpCAM+ CD45− cells. Rae1ε+ IECs are indicated by percentage. Two independent experiments (n=5 mice per group); mean; ANOVA / Tukey’s multiple comparison.