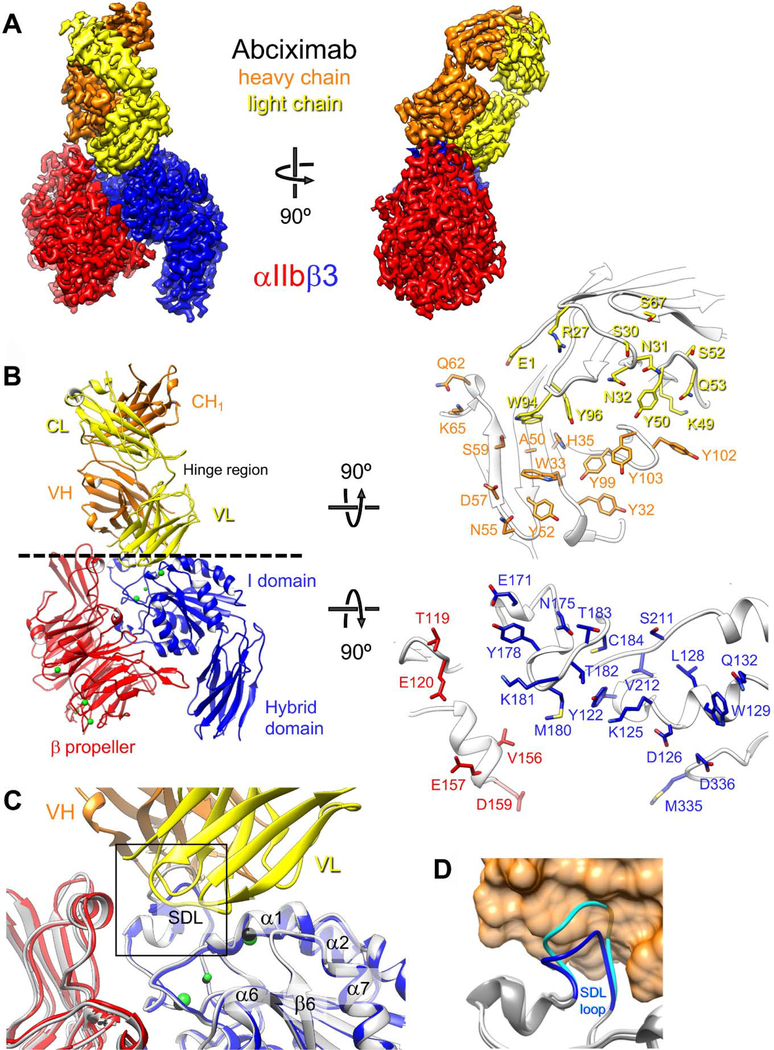
Figure 1.
Cryo-EM structure of the αIIbβ3–abciximab complex. (a) Density map of the αIIbβ3–abciximab complex at 2.8-Å resolution, segmented and colored according to the individual polypeptide chains. (b) Left panel: Atomic model of the αIIbβ3–abciximab complex colored as in panel (a). The green spheres represent the metal ions in the MIDAS, ADMIDAS and SyMBS. VH: variable domain of the heavy chain; CH1: first constant domain of the heavy chain; VL: variable domain of the light chain; CL: constant domain of the light chain. Hinge region denotes the flexible linkers that connect the variable and constant domains in abciximab. Right panel: Interaction surfaces of abciximab (top) and αIIbβ3 (bottom). Side chains of residues that are within 4.5 Å from a side chain of the interacting protein are shown in stick representation. (c) Fibrinogen/RGD-binding pocket in the cryo-EM structure of the αIIbβ3–abciximab complex (colored as in panel (a)) and in the X-ray crystal structure of the unliganded αIIbβ3 integrin (light grey with metal ions shown as black spheres). The overlay shows that abciximab binding does not induce meaningful differences in the binding pocket. The boxed area is shown in panel d. (d) Conformation of the β3 SDL loop in the cryo-EM structure of the αIIbβ3–abciximab complex (dark grey with SDL in blue) and in the X-ray crystal structure of the unliganded αIIbβ3 integrin (light grey with SDL in cyan). Abciximab is shown as an orange transparent surface to illustrate that the SDL in the conformation seen in the X-ray crystal structure of the unliganded αIIbβ3 integrin (cyan) would clash with abciximab.