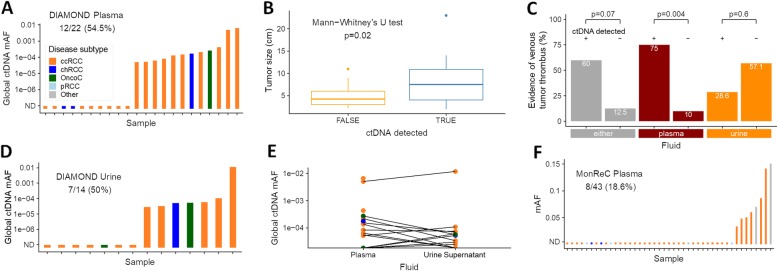
Fig. 3.
ctDNA detection using targeted assays. a Application of INVAR-TAPAS to DIAMOND plasma samples. ctDNA was detected in plasma of 12/22 (54.5%) patients, with global ctDNA mAF (gmAF) shown on the y-axis. Disease subtype is indicated by bar color, see insert for figure legend. b Assessment of the correlation between primary tumor size (diameter, cm), and ctDNA detection. Detection was via tMAD and/or INVAR-TAPAS, and in either fluid. This observation was driven by plasma (Additional file 1: Fig. S11A) with no apparent relationship in urine (Additional file 1: Fig. S11B). c ctDNA detection in plasma by INVAR-TAPAS was significantly more frequent amongst patients with venous tumor thrombus as compared to those without. This was not the case when considering ctDNA in urine or ctDNA in either fluid (Additional file 1: Fig. S12A-C). d INVAR-TAPAS was applied to DIAMOND USN samples. ctDNA was detected in 7/14 (50%) patients. e Comparison of gmAF of plasma and USN samples. In patients for whom we had access to both fluids, lines connect data points (Spearman’s rho = 0.28, p = 0.3). f Summary of targeted sequence analysis using a 10-gene QIASeq panel. Mutations at baseline were detected in 8/43 (18.6%) MonReC plasma samples. The y-axis denotes mAF which ranged from 3.5 × 10−2–0.15 (if two or more mutations were detected, the mean was calculated)