Abstract
Anthracnose is one of the major problems for cultivating many crops, including vegetables, fruits, and trees. It is a continual threat for fruits grower worldwide. Colletotrichum fructicola was isolated from Shine Muscat berries showing typical anthracnose symptom in Korea. It was identified as C. fructicola based on morphology, pathological signs and concatenated sequences of internal transcribed spacer region of rDNA, glyceraldehyde-3-phosphate dehydrogenase, β-tubulin-2, chitin synthase-1, calmodulin, and the Apn2-Mat1-2 intergenic spacer and partial mating type (Mat1-2) gene. To the best of our knowledge, this is the first report first report of anthracnose of Shine Muscat caused by C. fructicola in Korea.
Keywords: Shine Muscat, anthracnose, Colletotrichum fructicola, Koch’s postulates
Shine Muscat (Vitis vinifera L.) is the grape cultivar breeded by crossing Akitsu-21 (Vitis labruscana Baily × V. vinifera) and “Hakunan” (V. vinifera) [1]. Ever since its introduction to South Korea, it has become popular for its sweet seedless large grapes, and unique mango flavor. Grape growers have replaced their traditional Campbell cultivar with Shine Muscat and therefore the cultivation area for Shine Muscat has been growing rapidly in South Korea [2].
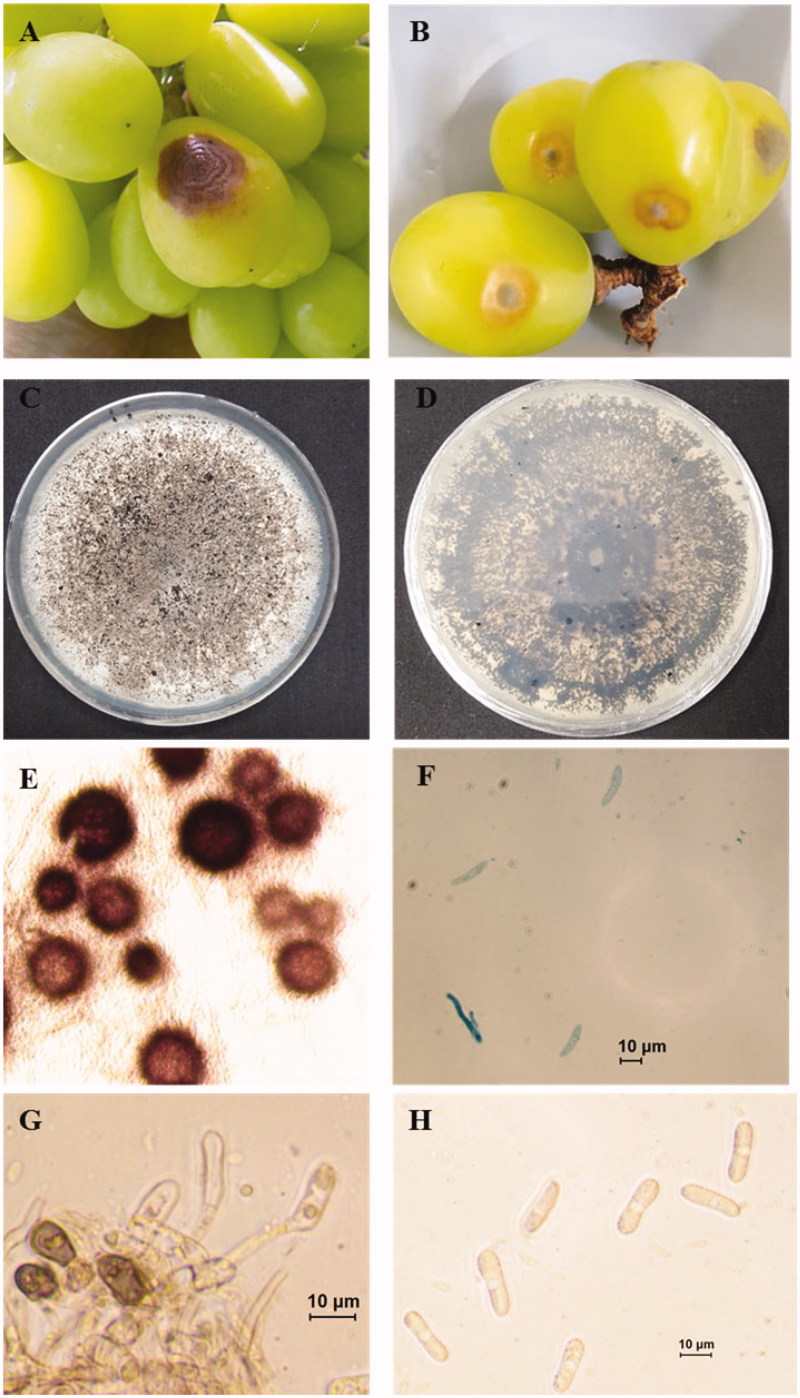
Anthracnose is a group of diseases caused by fungi especially Colletotrichum spp., that affects many plants, including vegetables, fruits, and trees. It is destructive to fruits, causing pre- and post-harvest decay. In August 2018, outbreak of anthracnose on Shine Muscat fruits occurred in a commercial orchard, Daegu, Korea and the disease incidence was approximately 25%. Symptoms were observed on the surface of fruits in the form of brown sunken lesion with concentric rings (Figure 1(A,B)). For the isolation of the causal agents, infected fruits showing typical anthracnose symptom brought to laboratory. Small sections containing healthy and necrotic tissues were cut from infected fruits, disinfested by immersing in 0.5% NaOCl for 3 min. The disinfested section of tissue rinsed in sterile water, dried by blotting, placed on water agar amended with 250 ppm streptomycin and incubated at 25 °C in the dark. After 3 days of incubation, fungal hyphae were emerged from the margin of infected tissue. Five-millimeter diameter plug of newly emerging hyphae was transferred to potato dextrose agar (PDA) and incubated at 25 °C in the dark. Pure cultures were obtained by following single spore isolation technique [3]. Primary conidial suspension was prepared in sterilized distilled water from 18 days old previously isolated colony. Concentration of conidial suspension was estimated using hemocytometer. Then 1 × 104 conidial suspension was prepared from the primary conidial suspension, sprayed over fresh PDA pates using sterilized wire loop and incubated at 25 °C in the dark. After 3 days, a single germinating conidium was transferred to new PDA plates, incubated at 25 °C in the dark and morphology of the colony was examined. Based on the similarity of the colony morphology, three isolates (ICKG2, ICKG4, and ICKG6) were selected for further morphological and molecular characterization. Twenty-day-old cultures were gray to white with immersing black perithecia in PDA (Figure 1(C,D)). For microscopic observation of teleomorph, 3–4 perithecia were picked up and mounted with lactophenol cotton blue on a glass slide. The sample was crushed by hitting gently over cover slide. Perithecia were brown and globose (Figure 1(E)). Ascospores were hyaline, slightly curved with round ends with the dimension of 13.5–25.6 µm × 4.0–6.1 µm (mean ± SD = 19.2 ± 2.9 × 5.0 ± 0.64 µm) (n = 25) (Figure 1(F)). Small mycelial mass was mounted on glass slide in sterilized distilled water for morphological analysis of morph. Abundant conidia were produced on short conidiophore. The conidia were hyaline, cylindrical, straight with rounded ends and 12.1–21.1 µm × 5.5–8.2 µm (mean ± SD = 15.9 ± 2.6 × 6.7 ± 0.9 µm) (n = 50) (Figure 1(G,H)). The glass slide containing conidia were put in a petri plate with moist tissue paper and incubated at 25 °C in the dark for enhancing appressoria formation. Appressoria were brown, various in shape with smooth edge and 9.4–13.7.1 µm × 5.9–8.2 µm (mean ± SD = 10.8 ± 1.3 × 7.1 ± 0.7 µm) (n = 20) (Figure 1(G)). The morphological characteristics of present isolates were match with those of previous described Colletotrichum species within C. gloeosporioides complex including C. fructicola [4–7]. Morphological characteristics of present isolates and close C. fructicola isolates are described in Table 1. Slight difference in morphological characteristics can be explained by growth media and type of host. Morphological characteristics of same species vary with the culture media and host type [4,8].
Figure 1.
Anthracnose of Shine Muscat caused by Colletotrichum fructicola. (A) Symptomatic Shine Muscat berries in infected orchard; (B) Sign and symptom appear on inoculated berries; (C, D) 20 days old fungal culture on PDA; (E) Perithecium; (F) Ascospore; (G) Conidiophore and appressoria (scale bar = 10 μm); (H) Conidia (scale bar = 10 μm).
Table 1.
Comparison of morphological characteristics between present isolate and previously reported Colletotrichum fructicola.
| Characteristics | Present isolate (ICK G4) | Colletotrichum fructicolaa | |
|---|---|---|---|
| Ascospores | Shape | Slightly curved with round ends | Fusiform-to-slightly curved with rounded ends |
| Color | Hyaline | Hyaline | |
| Size (μm) | 13.5 ∼ 25.6 × 4.0 ∼ 6.1 | 13.6 ∼ 24.0 × 2.6 ∼ 6.2 | |
| Conidia | Shape | Cylindrical, straight with rounded ends | Cylindrical, straight with rounded ends |
| Color | Hyaline | Hyaline | |
| Size (μm) | 12.1 ∼ 21.1 × 5.5 ∼ 8.2 | 13.1 ∼ 19.8 × 3.1 ∼ 7.0 | |
| Appressoria | Shape | Various in shape with smooth edge | Oval to fusoid with smooth edges |
| Color | Brown | Brown | |
| Size (μm) | 9.4 ∼ 13.7.1 × 5.9 ∼ 8.2 | – | |
aDescription and illustration [6].
Molecular analysis could be the best way to support morphological analysis. For achieving this objective, HiGene Genomic DNA Prep Kit (Biofact, Daejeon, South Korea) was used to extract total genomic DNA from selected isolates following manufacturer instructions. Internal transcribed spacer (ITS) region of rDNA and partial glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-tubulin-2 (TUB2), chitin synthase-1 (CHS-1), calmodulin (CAL), and the Apn2-Mat1-2 intergenic spacer and partial mating type (ApMat) genes were amplified using primer sets ITS1F and ITS4 [9,10], GDF and GDR [11], BT2a and BT2b [12], CHS-79F and CHS-345R [13], CL1C and CL2C [4], and AM-F and AM-R [14], respectively. Resulting PCR products were purified, sequenced through Macrogen Inc. (Seoul, Korea). The obtained sequences of each gene region were subjected to sequence similarity analysis using Basic Local Alignment Search Tool (BLAST) and deposited in GenBank [15]. The accession number and BLAST analysis result of each gene of present isolates is shown in Table 2. BLAST search result revealed that present isolates were 99-100% molecularly similar with the C. fructicola isolates (Table 2).
Table 2.
Molecular similarity of present isolates and previously reported Colletotrichum fructicola isolates.
| Present isolates | Gene (accession number) | Most similar Colletotrichum spp. isolates |
|---|---|---|
| ICKG2 | ITS (LC469121) | C. fructicola isolate ZJ-21 (100%) |
| GAPDH (LC469127) | C. fructicola isolate CPC 25976(100%) | |
| TUB2 (LC469124) | C. fructicola isolate HNLY-18 (99.76%) | |
| CHS-1 (LC469130) | C. fructicola isolate HJH-11 (100%) | |
| CAL (LC469133) | C. fructicola isolate HJH-11 (100%) | |
| ApMat (LC469136) | C. fructicola isolate CDTZG (100%) | |
| ICKG4 | ITS (LC469122) | C. fructicola isolate ZJ-21 (99.47%) |
| GAPDH (LC469128) | C. fructicola isolate CPC 25976(100%) | |
| TUB2 (LC469125) | C. fructicola isolate HNLY-18 (99.53%) | |
| CHS-1 (LC469131) | C. fructicola isolate HJH-11 (100%) | |
| CAL (LC469134) | C. fructicola isolate HJH-11 (99.86%) | |
| ApMat (LC469137) | C. fructicola isolate CDTZG (100%) | |
| ICKG6 | ITS (LC469123) | C. fructicola isolate ZJ-21 (99.16%) |
| GAPDH (LC469129) | C. fructicola isolate CPC 25976(100%) | |
| TUB2 (LC469126) | C. fructicola isolate HNLY-18 (100%) | |
| CHS-1(LC469132) | C. fructicola isolate HJH-11 (100%) | |
| CAL (LC469135) | C. fructicola isolate HJH-11 (100%) | |
| ApMat (LC469138) | C. fructicola isolate CDTZG (100%) |
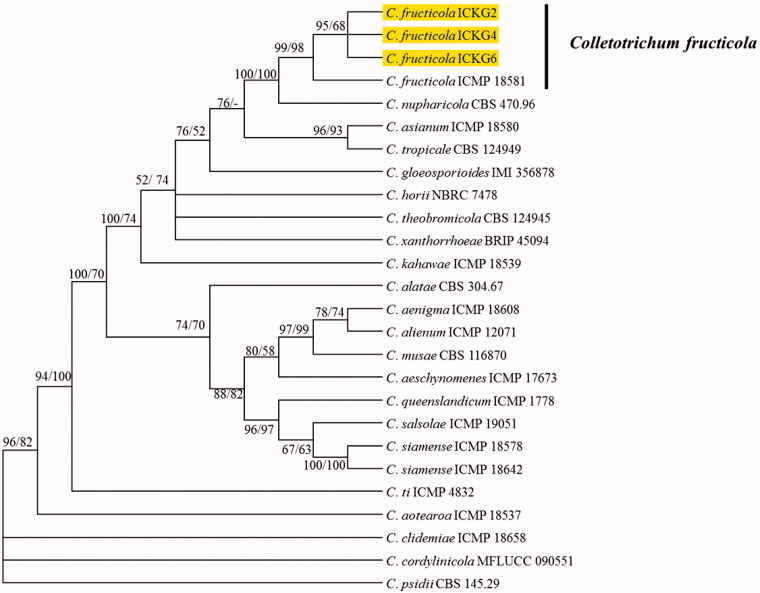
For phylogenetic analysis, individual gene sequences of present isolates and fungal species within C. gloeosporioides complex were aligned with MEGA v. 6.0 using MUSCLE program and then concatenated with Mesquite v. 2.75 [16,17]. Two phylogenetic trees were constructed using maximum likelihood and maximum parsimony statistical method from concatenated sequences (ITS, GAPDH, TUB2, CHS-1, CAL, and ApMat) in MEGA 6 [16]. The phylogenetic analysis delineated the present isolates from shine mascut as C. fructicola (Figure 2). Koch’s postulates were performed by inoculating healthy V. vinifera fruits with conidial suspension (1 × 106 conidia/ml) of ICKG2 isolate using wounding and unwounding methods. Seven-day-old culture of fungal isolate ICKG2 was used for preparing the conidial suspension. Fresh and heathy berries were collected from uninfected shine mascut orchard, surface sterilized with 0.5% NaOCl for 3 min followed by rinsing 2 times with sterile distilled water. The sterilized berries were dried by blotting and prepared for inoculation. For wounding method, berries were wounded with sterile pin to 1-mm depth and insulated with 10 µL droplet of conidial suspension. For un-wounding method, berries were sprayed with conidial suspension. The experiment was replicated 15 times (one fruits per replicate). Fruits received sterile water served as control. Treated fruits were incubated at 25 °C in the dark in a box with moist paper tissue. Anthracnose symptoms appeared on fruits (Figure 1(B)), inoculated by wounding method after 7 days and disease incidence was (80%). Only 30% of inoculated fruits with un-wounding method showed symptom after 14 days, whereas control fruits remained asymptotic. Same fungus (C. fructicola) was reisolated from inoculated fruits and identified following the methods described above. Based on morphology, molecular characterization, and pathological signs, present isolates were identified as C. fructicola.
Figure 2.
The phylogenetic tree generated by a maximum parsimony analysis of a combined dataset of ITS, GAPDH, TUB2, CHS-1, CAL, and ApMat gene sequences of Colletotrichum fructicola and those of other Colletotrichum spp. obtained from GenBank. Maximum persimony (MP) and maximum likelihood (ML) bootstrap support values (MP/ML ≥ 50%) are given at the corresponding node. The present isolate is indicated by yellow color.
C. fructicola is a representative fungal species with in Musae clade of C. gloeosporioides complex [4]. It was isolated from coffee berry and described as a pathogenic fugal species by Prihastuti et al. for the first time [5]. Now, it is known as the dominant and aggressive fungal species responsible for anthracnose on many crops including apple, sandy pear (Pyrus pyrifolia), strawberry, and avocado [6,7,18,19]. To the best of our knowledge, there were no report on anthracnose of Shine Muscat caused by C. fructicola in Korea, this is the first such study. The outcome of this will help to choose the right control measure against anthracnose of Shine Muscat, the increasingly popular grape cultivar in South Korea.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Yamada M, Yamane H, Sato A, et al. . New grape cultivar ‘Shine Muscat’. Bull Natl Inst Fruit Tree Sci. 2008;7:21–38. [Google Scholar]
- 2.KREI 2018 Korea Rural Economic Institute. Available from: http://www.krei.re.kr/eng/index.do
- 3.Cai L, Hyde KD, Taylor PWJ, et al. . A polyphasic approach for studying Colletotrichum. Fungal Divers. 2009;39:183–204. [Google Scholar]
- 4.Weir BS, Johnston PR, Damm U. The Colletotrichum gloeosporioides species complex. Stud Mycol. 2012;73:115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prihastuti H, Cai L, Chen H, et al. . Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 2009;39:89. [Google Scholar]
- 6.Zhang PF, Zhai LF, Zhang XK, et al. . Characterization of Colletotrichum fructicola, a new causal agent of leaf black spot disease of sandy pear (Pyrus pyrifolia). Eur J Plant Pathol. 2015;143(4):651–662. [Google Scholar]
- 7.Kim C, Hassan O, Lee D, et al. . First report of anthracnose of Apple caused by Colletotrichum fructicola in Korea. Plant Dis. 2018;102:2653. [DOI] [PubMed] [Google Scholar]
- 8.Hassan O, Jeon JY, Chang T, et al. . Molecular and morphological characterization of Colletotrichum species in the Colletotrichum gloeosporioides complex associated with persimmon anthracnose in South Korea. Plant Dis. 2018;102:1015–1024. [DOI] [PubMed] [Google Scholar]
- 9.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. [DOI] [PubMed] [Google Scholar]
- 10.White TJ, Bruns T, Lee S, et al. . Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand DH, Sninsky JJ, et al., editors. PCR protocols a guide to methods and applications. New York: Academic Press: 1990. p. 315–322. [Google Scholar]
- 11.Templeton MD, Rikkerink EH, Solon SL, et al. . Cloning and molecular characterization of the glyceraldehyde-3-phosphate dehydrogenase-encoding gene and cDNA from the plant pathogenic fungus Glomerella cingulata. Gene. 1992;122:225–230. [DOI] [PubMed] [Google Scholar]
- 12.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carbone I, Kohn LM. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. [Google Scholar]
- 14.Silva DN, Talhinhas P, Várzea V, et al. . Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia. 2012;104:396–409. [DOI] [PubMed] [Google Scholar]
- 15.National Center for Biotechnology Information Genebank overview [Internet]. Bethesda (MD): National Center for Biomedical Information; 2009. [cited 2009 Nov 20]. Available from: http://www.ncbi.nlm.nih.gov/Genbank
- 16.Tamura K, Stecher G, Peterson D, et al. . MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maddison WP, Maddison DR Mesquite: a modular system for evolutionary analysis. Version 2.75. http://mesquiteproject.org. 2011.
- 18.Fuentes-Aragón D, Juárez-Vázquez SB, Vargas-Hernández M, et al. . Colletotrichum fructicola, a member of Colletotrichum gloeosporioides sensu lato, is the causal agent of anthracnose and soft rot in avocado fruits cv. “Hass”. Mycobiology. 2018;46:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan P, Nakata N, Suzuki T, et al. . Markers to differentiate species of anthracnose fungi identify Colletotrichum fructicola as the predominant virulent species in strawberry plants in Chiba Prefecture of Japan. J Gen Plant Pathol. 2017;83:14–22. [Google Scholar]