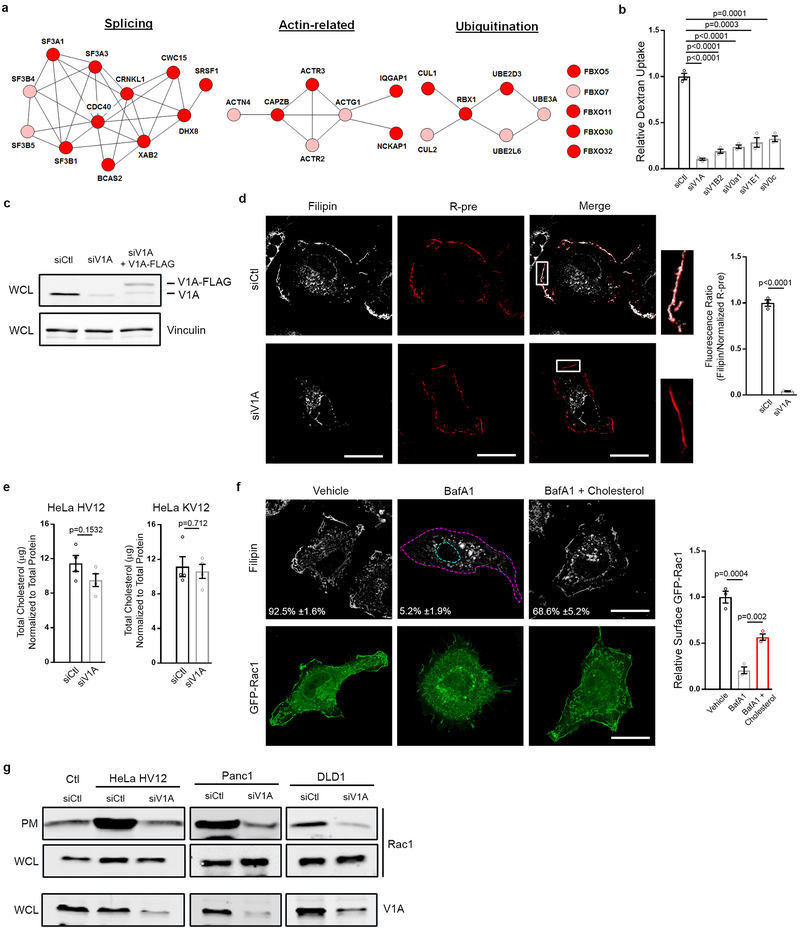
Extended Data Figure 1 |. v-ATPase is required for Ras-induced macropinocytosis.
a, Functional clusters within the macropinocytosis screen hits defined by STRING analysis (Pink, 1° screen; Red, 1° and confirmation screen). b, Quantification of TMR-dextran uptake following knockdown of the indicated v-ATPase subunits in HeLa HV12 cells. c, Immunoblot of V1A expression from whole cell lysate (WCL) (vinculin, loading control). d, Effect of v-ATPase depletion (siV1A) on cholesterol localization in Hela HV12 cells. Fluorescence micrographs of filipin staining (left), membrane labeling with R-pre (a transfected construct containing a modified sequence of the membrane targeting domain of KRas linked to RFP, middle), merge of filipin and R-pre with boxed areas enlarged to show PM localization (right), and the quantification of the ratio of filipin to R-pre membrane localization (bar graph) in control (siCtl) or V1A knockdown conditions. e, Effect of V1A knockdown on total cholesterol in HeLa HV12 (left) and KV12 (right) cells. f, Effect of Bafilomycin A1 (BafA1) and rescue by exogenous cholesterol on the localization of cholesterol and Rac1 in HeLa HV12 cells. Fluorescence micrographs of filipin (top), GFP-Rac1 (bottom) and quantification of relative surface GFP-Rac1 (bar graph). g, Effect of oncogenic Ras and V1A expression on Rac1 localization. Immunoblots of Rac1 and V1A in the plasma membrane (PM) fraction and whole cell lysate (WCL) from HeLa T7-vector control (Ctl) and HV12 or oncogenic KRas cell lines with or without V1A knockdown. Images (d,f) and immunoblots (c,g) are representative of three biological replicates. Dashed lines delineate the cell and nucleus and the mean percentages ±s.e.m shown represent the fraction of cells that display PM localization of cholesterol. Scale bars, 10μm. ≥500 (b,f) and 20 (d) cells were quantified in biological replicates (n=3). For e, cholesterol quantification is representative of four biological replicates. For all graphs, error bars indicate mean ±s.e.m. for indicated sample size, p value: unpaired, two-tailed Student t-test. For c,g, gel source data located in Supplementary Figure 1.