Figure 5. Identification of differential H2AZ.1 and H2AZ.2 interactors.
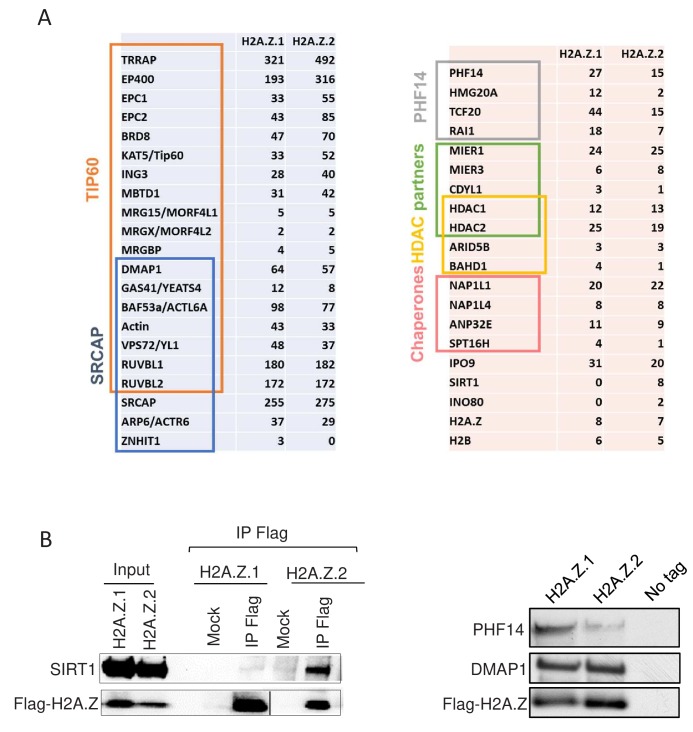
(A) Comparison of expression levels of tagged endogenous H2A.Z.1 (heterozygous) and H2A.Z.2 (homozygous) clones used in tandem affinity purification from nuclear extracts (see Figure 5—figure supplement 1). (B) Silver stained gel of fractions obtained for the purification of H2A.Z.1 and H2A.Z.2 from nuclear extracts of K562 cells shown in (A). A mock non-tagged cell line is used as control. Flag peptide elution is obtained from the first purification step (M2-Flag resin) and biotin elution from the second final step (Strep-Tactin resin). Known components of protein complexes interacting with histone H2A.Z are identified on the right (PHF14 is also indicated). (C) Dot-blot representation of AP-MS experiments using tagged H2A.Z.1 and H2A.Z.2 as baits. Circle filling represents average spectral counts, while circle diameter represents relative enrichment in one bait versus the other and circle border represents BFDR confidence level. Known/expected partners based on the literature and large-scale public data (BioGrid) include TIP60/p400, SRCAP and HDAC complexes. Data represent two replicates for each bait and were normalized on H2AZ-H2B chaperone levels (ANP32E, NAP1L1 and NAP1L4). (D) Western-blot validation of interactions shown in (B–C). TAP-purified fractions were normalized based on Flag-H2A.Z signals, loaded on SDS-PAGE gels, and blotted with the indicated antibodies.