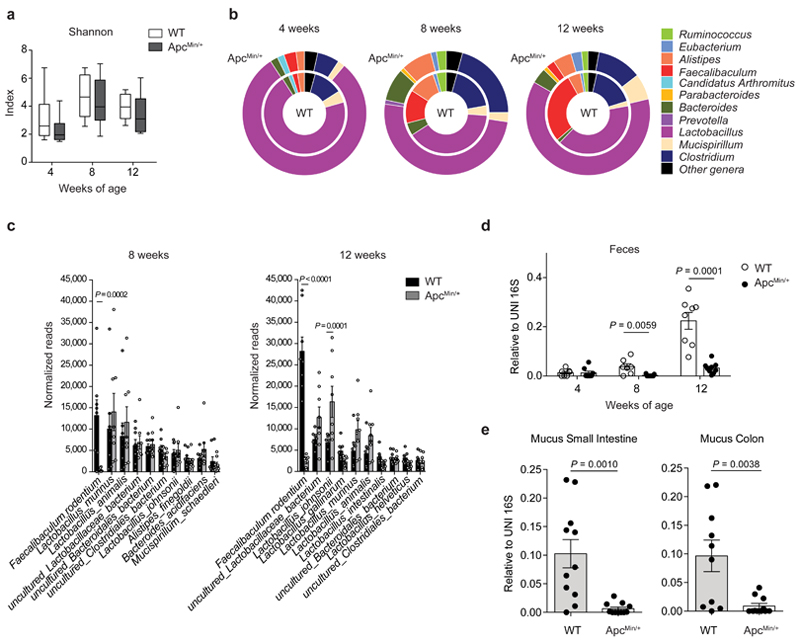
Fig. 1. Faecalibaculum rodentium is underrepresented during the early phases of tumor development.
a-c,16S rRNA gene profiling of the fecal microbiota of WT and ApcMin/+ mice at 4, 8 and 12 weeks of age (n = 8 mice/group). a, Shannon diversity index. Box plots show the interquartile range, median value and whiskers min to max. b, Genus abundance (inner pie: WT, outer pie: ApcMin/+). Genera with a relative abundance higher than 1% in at least one of the tested condition, were shown, otherwise are collapsed into the “Other genera” section. P values were assessed by two-tailed unpaired Mann-Whitney test. c, Relative abundance of the 10 most abundant species in fecal bacterial DNA isolated from WT and ApcMin/+ mice at 8 and 12 weeks of age. Abundance shown as the normalized number of assigned sequences in the 16S rRNA sequencing. P values were determined by two-way ANOVA with Bonferroni post-test. d,e, qPCR of F. PB1 abundance normalized to panbacterial primers targeting the 16S rRNA gene (UNI 16S) in bacterial DNA extracted from feces (d, n = 8 mice/group) and mucus from the small intestine and colon (e, n = 11 mice/group) from WT and ApcMin/+ mice. P values were determined by multiple t-tests corrected for multiple comparisons using the Holm-Sidak method to compare F. PB1 abundance between groups at each time point (d) or by two-tailed unpaired t-test (e). a-e, Data from two independent experiments and represented as means ± s.e.m.