Abstract
Background
The tumor microenvironment can be classified into immunologically active “inflamed” tumors and inactive “non-inflamed” tumors based on the infiltration of cytotoxic immune cells. Previous studies on liver cancer have reported a superior prognosis for inflamed tumors compared to non-inflamed tumors. However, liver cancer is highly heterogeneous immunologically and genetically, and a finer classification of the liver cancer microenvironment may improve our understanding of its immunological diversity and response to immune therapy.
Methods
We characterized the immune gene signatures of 234 primary liver cancers, mainly virus-related, from a Japanese population using RNA-Seq of tumors and matched non-tumorous hepatitis livers. We then compared them with the somatic alterations detected using the whole-genome sequencing.
Findings
Liver cancers expressed lower levels of immune marker genes than non-tumorous hepatitis livers, indicating immunosuppression in the tumor microenvironment. Several immunosuppression mechanisms functioned actively and mutually exclusively, resulting in four immune subclasses of liver cancer: tumor-associated macrophage (TAM), CTNNB1, cytolytic activity (CYT), and regulatory T cell (Treg). The CYT and Treg subclasses represented inflamed tumors, while the TAM and CTNNB1 subclasses represented non-inflamed tumors. The TAM subclass, which comprised 31% of liver cancers, showed a poor survival, expressed elevated levels of extracellular matrix genes, and was associated with somatic mutations of chromatin regulator ARID2. The results of cell line experiments suggested a functional link between ARID2 and chemokine production by liver cancer cells.
Interpretation
Primary liver cancer was classified into four subclasses based on mutually exclusive mechanisms for immunosuppression. This classification indicate the importance of immunosuppression mechanisms, such as TAM and Treg, as therapeutic targets for liver cancer.
Funding
The Japan Agency for Medical Research and Development (AMED).
Keywords: Liver cancer, Tumor microenvironment, Tumor-associated macrophage, Regulatory T cell
Abbreviations: TAM, tumor-associated macrophage; Treg, regulatory T cell; TME, tumor microenvironment; HCV, hepatitis C virus; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; ICC, intrahepatic cholangiocarcinoma; cHCC-ICC, combined hepatocellular carcinoma-intrahepatic cholangiocarcinoma; WGS, whole genome sequencing; ICGC, International Cancer Genome Consortium; FPKM-UQ, fragments per kilobase of exon per million fragments mapped with upper quartile normalization; CYT, cytolytic activity; CNA, copy number alternation; PCAWG, Pan-Cancer Analysis of Whole Genomes; HR, hazard ratio; CI, confidence interval; OR, odds ratio; GSEA, gene set enrichment analysis; ECM, extracellular matrix; FDR, false discovery rate; SNV, single nucleotide variant; INDEL, insertion and deletion; TCGA, The Cancer Genome Atlas
Research in context.
Evidence before this study
Tumors are composed of both cancer cells and other types of cells, including fibroblasts, endothelial cells, and leukocytes. The composition of leukocytes in tumors is of particular interest due to its relation with the patient's prognosis and response to therapy. A previous study revealed that liver cancer can be classified into leukocyte-rich “inflamed” cancer and leukocyte-poor “non-inflamed” cancer, where patients with “inflamed” liver cancer have a better prognosis.
Added value of this study
Liver cancer can be classified into four immunological subclasses. Inflamed liver cancer can be subclassified into the “CYT” and “Treg” subclasses, while non-inflamed liver cancers are divided into the “TAM” and “CTNNB1” subclasses. The TAM and Treg subclasses are infiltrated by immune suppressive leukocytes and are associated with a poor prognosis.
Implications of all the available evidence
This study revealed the immunological heterogeneity of liver cancer. The immunological classification presented in this work may be used to guide patient stratification for immune checkpoint therapy.
Alt-text: Unlabelled box
1. Introduction
Tumor tissue not only contains tumor cells but also other cells, such as leukocytes, endothelial cells, and fibroblasts, which collectively constitute the tumor microenvironment (TME). Cells in the TME closely interact with tumor cells and exert both pro- and anti-tumor effects. The key players of the anti-tumor effect are immune cells, including cytotoxic T lymphocytes and natural killer (NK) cells. Based on the level of cytotoxic immune cells infiltrating into the TME, tumors can be classified as immunologically active “inflamed” tumors or immunologically inactive “non-inflamed” tumors. Inflamed tumors have a better prognosis than non-inflamed tumors in lung, colorectal, and ovarian cancers, among others [1], [2], [3], [4]. Furthermore, infiltration by cytotoxic immune cells can be used to predict responses to immune checkpoint inhibitors, albeit to a limited degree [5]. This demonstrates the clinical utility of this binary classification, while at the same time suggesting the need for a finer immunological classification of TME.
Liver cancer is the 4th leading cause of cancer-related death in the world. Liver cancer mostly develops from chronic hepatitis and is infiltrated by various immune cells. Its TME can also be classified into inflamed and non-inflamed tumors [6]. An inflamed TME of liver cancer is associated with a significantly better overall survival [7], [8], [9] and disease-free survival [10,11] compared to non-inflamed TME. In addition, some liver cancers appear to respond well to immune checkpoint inhibitors [12,13]. Whether or not the response correlates with inflamed TME is currently unclear. However, because the response was found to be correlated with PD-L1 expression [13], TME may be a key factor for the response of liver cancer to immune checkpoint inhibitors.
While the “inflamed or not” classification of liver cancer is useful, it may oversimplify and neglect the immunological complexity of clinical samples. For example, a single-cell RNA-Seq study revealed that diverse subpopulations of T cells infiltrate into liver cancer [14]. The ratio of Tregs to CD4+ or CD8+ T cells is high in some liver cancer tissues, and these cancers are associated with a poor prognosis [11,15]. The level of infiltrating CD68+ macrophages also differs among liver cancers, and has been shown to have a negative impact on the survival of patients [16]. This immunological heterogeneity among liver cancers requires a finer classification of the immunological microenvironment beyond inflamed/non-inflamed tumors. A more precise classification and its correlation with genomic alterations may be used to guide clinical decision-making for treatment with immuno-checkpoint inhibitors in cancer genomic medicine, as well as provide a basis for the development of novel therapeutic targets.
Here, we performed immunogenomic analyses using the transcriptome and whole-genome sequencing data on 234 liver cancers in a Japanese population [17]. The majority of the liver cancers were positive for hepatitis C virus (HCV) or hepatitis B virus (HBV) and thus etiologically distinct from the Western cohorts. We found that inflamed and non-inflamed liver cancers were each divided into two subclasses based on their immunosuppression mechanisms. The subclasses had unique associations with etiology, prognosis, molecular subtypes, and somatic alterations. Our results shed light on the heterogeneity of TME in clinical samples of liver cancer.
2. Materials and methods
2.1. RNA-Seq and whole genome sequencing for liver cancers
In a previous study, we performed whole genome sequencing (WGS) for 300 liver cancers and RNA-Seq for 259 of 300 tumors, as part of the International Cancer Genome Consortium (ICGC) [17]. In this study, we computationally re-analyzed 234 out of the 259 tumors. The data for the other 25 tumors was omitted because they were not submitted to mutation calling by the Pan-Cancer Analysis of Whole Genomes (PCAWG) project (see below) [18]. The RNA-Seq data of matched non-tumor liver tissues were available for 196 out of the 234 cases. The clinical information of the patients is summarized in Table 1. All patients provided written informed consent for their participation in the study following the ICGC guidelines. Institutional review boards at RIKEN and all groups participating in this study approved this work. We re-analyzed the RNA-Seq reads of liver cancers and non-tumorous livers. Read mapping onto reference human genome (GRCh37) with TopHat2, and the read counting for GENCODE release 19 with HTseq, were orchestrated by the iRAP pipeline [19]. Fragments per kilobase of exon per million fragments mapped with upper quartile normalization (FPKM-UQ) were computed and used as the gene expression levels throughout the study. When log expression was required, 0.01 was added to FPKM-UQ as offset.
Table 1.
Clinical information of the cohort.
| Number of patients | 234 |
|---|---|
| Age, median (Q1–Q3) | 68.5 (62–74) |
| Gender, male (%) | 174 (74) |
| Histology (%) | |
| HCC | 208 (89) |
| ICC | 19 (8) |
| cHCC-ICC | 7 (3) |
| Virus (%) | |
| HCV | 128 (55) |
| HBV | 58 (25) |
| NBNC | 44 (19) |
| HBV, HCV | 4 (2) |
| Tumor size, > 3 cm (%) | 113 (48) |
| Vascular invasion (%) | 80 (34) |
| Stage (%) | |
| I | 39 (17) |
| II | 106 (45) |
| III | 69 (30) |
| IV | 20 (9) |
| AFP, > 200 ng/mL (%) | 65 (28) |
| Platelet count, < 100,000/µL | 51 (22) |
| Five-year overall survival (95% CI) | 0.63 (0.56–0.70) |
| Five-year disease-free survival (95% CI) | 0.33 (0.27–0.40) |
2.2. Immune signature
Cytolytic activity (CYT) was computed as the geometric mean between the FPKM-UQ of PRF1 and GZMA. The gene expression signatures of CD8 T cells, NK cells, B cells, interferon-α response, interferon-γ response, and Wnt/β-catenin signaling were computed using the single-sample GSEA module on the GenePattern server (https://genepattern.broadinstitute.org/). The signature genes for the CD8 T cells, NK cells, and B cells were retrieved from a previous study [20]. As the signature gene sets for interferon responses and Wnt/β-catenin signaling, we downloaded “HALLMARK INTERFERON ALPHA RESPONSE,” “GO CELLULAR RESPONSE TO INTERFERON GAMMA,” and “CHIANG LIVER CANCER SUBCLASS CTNNB1 UP” from the Molecular Signatures Database (MSigDB, version 6.1). The member genes of these signatures are listed in Supplementary Table 1. The molecular subclasses of Chiang et al. [21] and Hoshida et al. [22] were assigned to our liver cancer samples by using the nearest template prediction on GenePattern. The signature genes for the molecular subclasses were obtained from MSigDB. The immune classification of liver cancer was performed in a similar manner, using a previously published gene set [9]. The ESTIMATE immune score and stromal score were computed using the ESTIMATE R package. The prediction of the immune cell fractions by CIBERSORT was performed by submitting FPKM-UQ to the public server (https://cibersort.stanford.edu).
2.3. Immunohistochemistry
From 70 formalin-fixed, paraffin-embedded tissue samples, which were analyzed previously by RNA-Seq and WGS in their frozen tissues [17], five 4-μm-thick sections were serially cut and mounted on pre-coated slides. A FOXP3 assay was then performed using the Ventana Benchmark XT system (Roche). For antigen retrieval, Cell Conditioning 1 (Roche) was poured onto the sections, which were then heated on a slide heater at 95 °C for 64 min. The tissue sections were incubated with × 200 dilution of FOXP3 antibody (Cell Signaling Technology). A CD163 assay was performed using DAKO Autostainer Link 48 (Agilent Technologies). For antigen retrieval, the sections were heated at 97 °C for 20 min with Target Retrieval Solution, High pH (Agilent Technologies). The tissue sections were incubated with × 200 dilution of CD163 antibody (Novocastra, Leica Biosystems).
2.4. Somatic mutation call from whole genome sequencing data
Somatic mutation calls for SNVs, indels, SVs, and copy number alternations (CNAs) were previously generated by the Pan-Cancer Analysis of Whole Genomes (PCAWG) project [18] as part of efforts to analyze 2800 pan-cancer WGS using uniform pipelines. The somatic SNVs and INDELs were called as a consensus of four individual variant callers. For the somatic CNA calls, a consensus of six individual CNA callers were used. These data is available online at https://dcc.icgc.org/pcawg.
2.5. Clustering of liver cancer
To immunologically classify the liver cancers, we selected four gene expression signatures: macrophages M2, Wnt/β-catenin signaling, Tregs, and cytolytic activity. The first three signatures represent known immunosuppression mechanisms, whereas the last one represents anti-tumor immunity. For Tregs and macrophage M2, we used the fraction of these cell types in tumor tissue estimated by CIBERSORT. For Wnt/β-catenin signaling, we computed a signature using single-sample GSEA with the genes listed in Supplementary Table 1. Cytolytic activity was transformed using logarithm. These four signatures were scaled across samples, and distances between samples were defined as 1 ‒ Pearson's correlation. The hierarchical clustering of the 243 samples was performed using the Ward linkage, and the dendrogram was cut into four clusters after visual inspection.
2.6. Microarray analysis and RNA-Seq of HCC cell lines
Two HCC cell lines, JHH4 and JHH5, were analyzed. The CRISPR/Cas9-mediated knockout of ARID2, cell culture, RNA extraction, and microarray profiling of these cells were performed as previously described [23]. The gene expression levels were quantified from CEL files using the robust multi-array average (RMA) method implemented in the Bioconductor “affy” package. Log-fold changes of gene expression between the knockout and wild-type cells were computed using the Bioconductor “limma” package. GSEA was performed by uploading the RMA expression measures to the GenePattern server. We also performed RNA-seq of the wild-type and knock-out JHH4 cells (triplicated) by using the TruSeq stranded mRNA library Prep kit (Illumina) and the data analysis was performed by the same method for the liver cancer samples.
2.7. Statistical analysis
All statistical tests were two-sided and performed using R software (version 3.6.0). The log-rank test for trend was performed using the “survMisc” package of R. For a single hypothesis testing, the significance threshold was p < 0.05. For multiple hypothesis testing, the significance threshold was a false discovery rate (FDR) < 0.05 after the Benjamin–Hochberg correction.
2.8. Data availability
WGS and RNA-Seq reads for matched tumor and non-tumor tissues are available in the European Genome-phenome Archive database (accession number EGAS00001000678). The gene expression data for cell lines were deposited to the NCBI's Gene Expression Omnibus. The accession number is GSE144021 for microarray and GSE143941 for RNA-Seq.
3. Results
3.1. Inflammation in liver cancer confers better prognosis
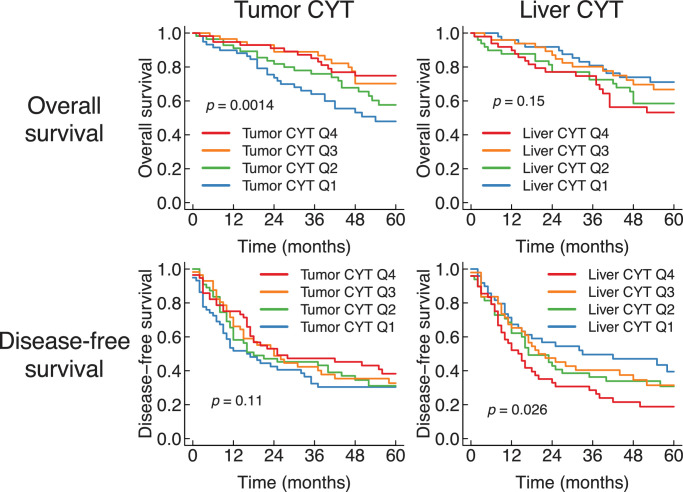
Previous studies have reported that inflamed TME in liver cancer is associated with a better prognosis [7], [8], [9], [10], [11]. To confirm this, we first compared the level of inflammation in liver tissues with the prognosis of patients. As a measure of inflammation, we examined cytolytic activity (CYT), which is defined as the average expression of granzyme A (GZMA) and perforin (PRF1) [24]. We computed CYT for both tumors and adjacent non-tumorous livers, and examined their effect on overall survival and disease-free survival (Fig. 1). Patients with high tumor CYT had a significantly better overall survival compared to low tumor CYT (p = 0.0014 by log-rank test for trend). The prognostic value of tumor CYT was independent from age, gender, and tumor stage (p = 0.002 by Cox proportional hazards model).
Fig. 1.
Inflammation of liver cancer and non-tumorous liver were oppositely correlated with prognosis. Kaplan–Meier plots of overall and disease-free survival after surgical treatment of liver cancer. The patients were stratified by the cytolytic activity of the tumor (Q1, N = 59; Q2, N = 56; Q3, N = 57; Q4, N = 57) or the liver (Q1, N = 49; Q2, N = 49; Q3, N = 49; Q4, N = 49). The p-values were computed using the log-rank test for trend.
We also compared other measures of inflammation with prognosis. The ESTIMATE immune score [25] and the immune classes proposed by Sia et al. [9] reflect broader aspects of immune response than CYT, taking 141 and 112 genes into consideration, respectively. Concordantly with tumor CYT, a high tumor ESTIMATE immune score was associated with a better overall survival (p = 0.033 by log-rank test for trend) (Supplementary Fig. 1a). The Sia immune class of tumor was not associated with overall survival (p = 0.75 by log-rank test) (Supplementary Fig. 1b). As CYT had the strongest correlation with prognosis, we used CYT as the main readout of inflammation in this study.
In contrast to tumor CYT, CYT in non-tumorous hepatitis livers was associated with adverse outcomes (Fig. 1). Patients with high CYT in adjacent non-tumorous liver had a poor disease-free survival compared to low CYT (p = 0.026 by log-rank test for trend). This unfavorable prognosis by non-tumorous liver inflammation could represent liver failure or multi-centric occurrence due to chronic hepatitis and cirrhosis [26].
A favorable prognosis based on tumor inflammation could be attributed to the activity of anti-tumor immunity. Since tumor inflammation provides a prognostic benefit, we investigated how the level of tumor inflammation is affected by clinical, pathological, and molecular factors.
3.2. Tumor microenvironment of liver cancer is immunosuppressive
To identify factors that influence tumor inflammation, we compared inflammation in hepatitis livers, where the majority of liver cancers originate from. Viral hepatitis is a major etiology of liver cancer in Asia [27]. We found that, although non-tumorous liver CYT had a significant association with the type of virus (p = 0.00015 by Kruskal–Wallis test), tumor CYT did not (p = 0.47) (Supplementary Fig. 2a). Similarly, tumor CYT was not correlated with the histological stage of liver fibrosis (Inuyama classification; p = 0.83) (Supplementary Fig. 2b). We also compared CYT in tumors with CYT in non-tumorous livers from matched patients. Although tumor and liver CYT was positively correlated, the degree of correlation was weak (Spearman correlation coefficient 0.15, p = 0.035) (Supplementary Fig. 2c). Taken together, tumor inflammation was poorly correlated with virus, liver fibrosis, and liver inflammation, indicating that TME is less influenced by background liver inflammation.
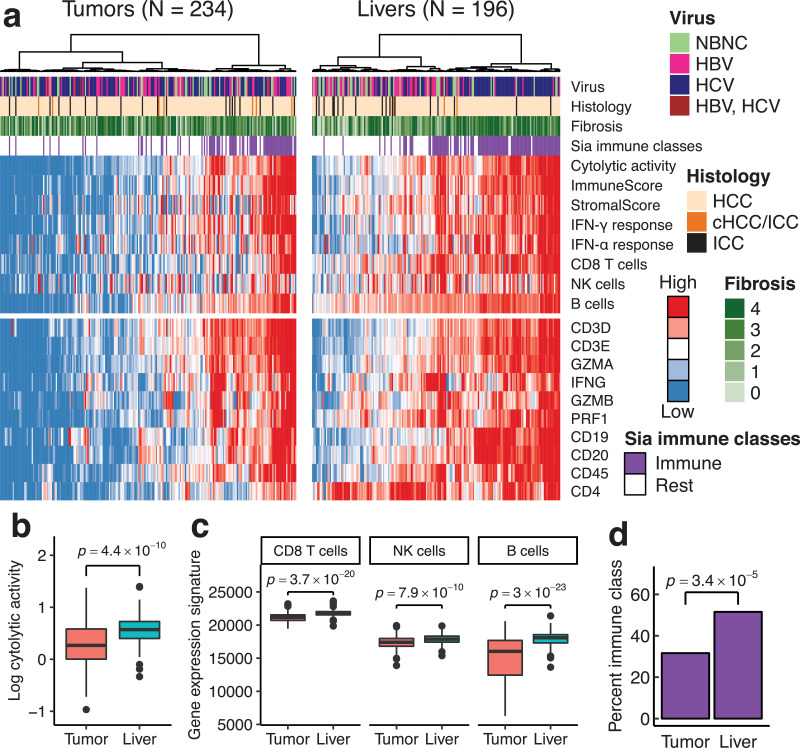
The comparison of CYT showed that tumors are less inflamed than non-tumorous liver (median CYT, 1.84 and 3.71, respectively; p = 4.4 × 10−10 by Wilcoxon signed-rank test) (Fig. 2b). To confirm this, we examined the expression levels of the representative inflammatory genes (Fig. 2a). Markers of T cells (CD3E, CD8A, and CD4), B cells (CD19 and CD20), and leukocytes (CD45) had significantly decreased expression levels in tumors compared to in non-tumor livers (p-values < 0.0001 by Wilcoxon signed-rank test). Likewise, interferon-γ (IFNG) and cytotoxic molecules (GZMA, GZMB, and PRF1) were less expressed in the tumors (p-values < 1.0 × 10−5 by Wilcoxon signed-rank test). The multigene signatures of the immune response were also reduced in the tumor. The ESTIMATE immune score was lower in the tumor than that in non-tumorous livers (median, 753 and 1370, respectively; p = 1.9 × 10−13 by Wilcoxon signed-rank test) (Supplementary Fig. 2d). The gene expression signatures of the CD8 T cells, NK cells, B cells, and interferon responses were diminished in the tumors (Fig. 2c; Supplementary Fig. 2e) (p-values < 1.0 × 10−9 by Wilcoxon signed-rank test). The increased expression of the B cell signature in non-tumorous livers may represent the formation of intrahepatic lymphoid follicles in chronic hepatitis C [28]. Consistently with these quantitative measures, the Sia immune class was less frequent in tumors (74 of 234, 32%) than in non-tumorous livers (101 of 196, 52%; p = 3.4 × 10−5 by Fisher's exact test) (Fig. 2d).
Fig. 2.
Liver cancers were less inflamed than non-tumorous livers. (a) The expression levels of immune-related genes and multigene signatures in tumor and non-tumorous liver. Samples are ordered by hierarchical clustering of gene expression levels. NBNC, negative for both HBV and HCV; HCC, hepatocellular carcinoma; ICC, intrahepatic cholangiocarcinoma; cHCC/ICC; combined HCC-ICC. (b, c) Immune gene expression signatures in matched tumor and non-tumorous liver. P-values were computed using Wilcoxon signed-rank test. (b) Cytolytic activity. (c) Immune cell signatures. (d) The fraction of Sia's immune class in tumor and non-tumorous liver.
These results demonstrated that tumors were immunologically quiescent compared to non-tumorous livers. Liver cancers that originated from an inflamed liver may have acquired the immunosuppressive phenotype during the course of liver carcinogenesis.
3.3. Immunosuppression subclasses of liver cancer
Several mechanisms have been proposed to explain the immunosuppression in liver cancer. Although the activation of WNT/β-catenin signaling is associated with the exclusion of T cells from tumor, little is known about its mechanism. This association was initially reported in melanoma [29], but similar trends have been observed in multiple cohorts of HCC [9,11,30]. Other proposed mechanisms include the infiltration of immunosuppressive leukocytes in liver cancer. Tregs and TAMs are abundant in liver cancer [14,31,32], and their infiltration is associated with a poor prognosis [15,16,33]. Consistent with the immunosuppressive role of TAM, the fraction of macrophages M2 had a strong negative correlation with CYT in our data set (Supplementary Fig. 3).
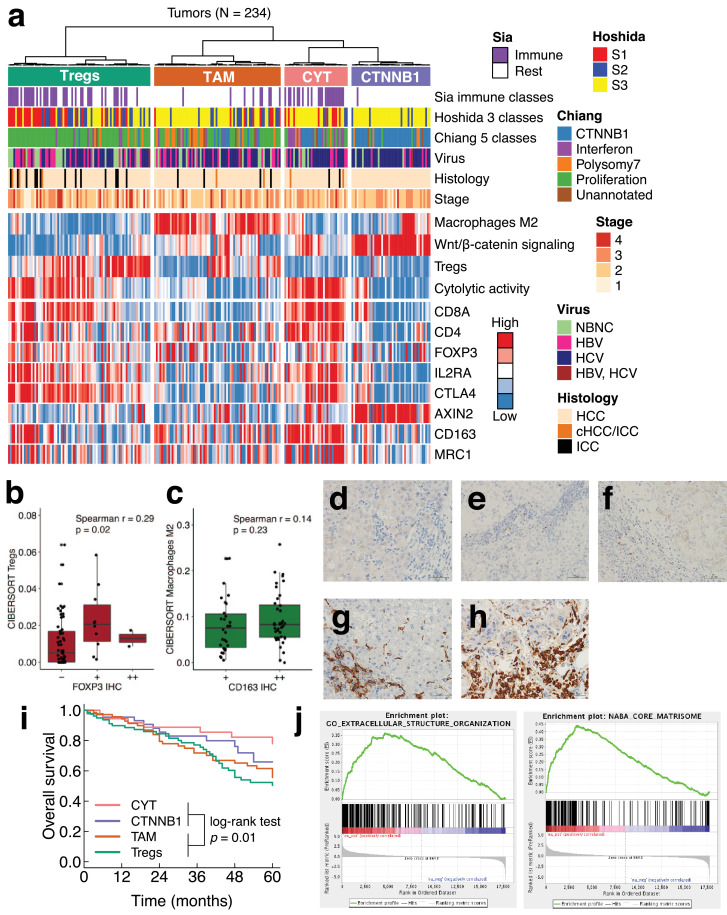
However, it remains unclear whether these immunosuppression mechanisms work jointly or separately in clinical samples of liver cancer, which are diverse in terms of pathology. To investigate this issue, we performed an unsupervised hierarchical clustering of liver cancer using four gene expression signatures (Fig. 3a). Among the four signatures, three signatures represented immunosuppressive components (Tregs, TAM, and WNT/β-catenin signaling), and one signature represented anti-tumor immunity (CYT). For the Tregs and TAM signatures, we estimated the tumor-infiltrating fractions of the cells using CIBERSORT [34]. The CIBERSORT estimates of Tregs and TAM were positively correlated with the immunohistochemistry of FOXP3 and CD163, respectively (Fig. 3b–h). For the WNT/β-catenin signaling signature, we used a previously defined gene set [21].
Fig. 3.
Classification of liver cancer based on immunosuppression mechanisms. (a) Unsupervised clustering of 243 liver cancers by four gene expression signatures related to immunity (macrophages M2, Wnt/β-catenin signaling, Tregs, cytolytic activity). Expression levels of marker genes for T cells (CD8A and CD4), Tregs (FOXP3, IL2RA, and CTLA4), Wnt/β-catenin signaling (AXIN2), and macrophages M2 (CD163 and MRC1) are also shown. (b, c) Comparison of infiltration levels between immunohistochemistry and the absolute mode of CIBERSORT. (b) Treg marker FOXP3 and CIBERSORT estimates. (c) Macrophage M2-marker CD163 and CIBERSORT estimates. (d–f) IHC of FOXP3. (d) FOXP3 ‒, (e), FOXP3 +, and (f) FOXP3 ++. (g, h) IHC of CD163. (g) CD163 +. (h) CD163 ++. (i) Patient overall survival and the immunosuppression subclasses. (j) Gene set enrichment plots. Upregulated genes in the TAM subclass compared to the CTNNB1 subclass are shown (FDR < 0.001).
As a result, 234 liver cancers were classified into four immune subclasses (Fig. 3a), namely the TAM, CTNNB1, CYT, and Treg subclasses, based on their characteristic immune signatures. The number of samples in these subclasses was 72, 45, 36, and 81 (31%, 19%, 15%, and 35%), respectively. The TAM and CTNNB1 subclasses represented non-inflamed tumors. Indeed, the TAM and CTNNB1 subclasses had a lower CYT (median CYT, 1.33 and 0.84, respectively) compared to the CYT and Treg subclasses (4.75 and 2.73; p = 4.1 × 10−21). Likewise, only 13% and 2% of the TAM and CTNNB1 subclasses were in Sia's immune class, whereas 64% and 51% of the CYT and Treg subclasses were in Sia's immune class, respectively (p = 9.7 × 10−15). The expression of immune checkpoint genes (e.g. PD1 and PD-L1) was highest in the CYT subclass, and second highest in the Treg subclass (Supplementary Fig. 4).
Our immunosuppression subclasses showed some parallels with the gene expression-based classification (Fig. 3a). A comparison with Chiang's classification [21] showed that our TAM subclass was overrepresented by Chiang's polysomy 7 class (odds ratio (OR) 3.55; p = 0.002). In addition, our CTNNB1, CYT, and Treg subclasses were overrepresented by Chiang's CTNNB1 (OR 31.2; p = 2.3 × 10−19), interferon (OR 9.45; p = 2.6 × 10−6), and proliferation classes (OR 10.0; p = 6.5 × 10−14), respectively. In terms of Hoshida's classification [22], our Treg subclass was overrepresented by Hoshida's S1 (OR 4.92; p = 2.3 × 10−6) and S2 subclasses (OR 1.96; p = 0.045). Our CTNNB1 subclass was overrepresented by Hoshida's S3 subclass (OR 6.26; p = 7.7 × 10−6).
Regarding etiology, HCV was overrepresented in the CTNNB1 subclass (OR 3.6; p = 0.0007) and underrepresented in the Treg subclass (OR 0.46; p = 0.006). HBV was underrepresented in the CTNNB1 subclass (OR 0.32; p = 0.02). In terms of patient prognosis, the TAM and Treg subclasses had a significantly poor overall survival compared to the CYT and CTNNB1 subclasses (p = 0.01 by log-rank test) (Fig. 3i). This difference was still significant even after correction for age, gender, and tumor stage (hazard ratio (HR) 1.89; 95% confidence interval (CI) 1.11–3.20).
Since non-inflamed tumors are often resistant to immune checkpoint therapy, the molecular characterization of these tumors may have clinical implications. Of the two non-inflamed subclasses, the CTNNB1 subclass was well-recognized, while the TAM subclass has not yet been characterized. To better understand the TAM subclass, we performed gene set enrichment analysis (GSEA) of the TAM subclass and found that significantly more genes were expressed related to the extracellular matrix (ECM) than in the CTNNB1 subclass (FDR < 0.001), suggesting the enhanced deposition and remodeling of the ECM (Fig. 3j). TAM secretes TGFB1, which induces ECM remodeling and immunosuppression [35]. Consistent with this, the late TGFB1 signature [36] was expressed more highly in the TAM subclass than the CTNNB1 subclass (FDR < 0.01) (Supplementary Fig. 5).
To determine whether the same classification applies for other cohorts, we analyzed the RNA-Seq of 193 HCC in The Cancer Genome Atlas (TCGA) project [6] (Supplementary Fig. 6). The TCGA cohort was etiologically distinct from our Japanese cohort. Patients negative for both HBV and HCV made up 61% of TCGA but 19% of the Japanese cohort. However, the TCGA liver cancers could be classified into the four subclasses (CTNNB1, 25%; CYT, 23%; TAM, 19%; Tregs, 33%), indicating the stability of our immunological classification.
Our analysis demonstrated that inflamed and non-inflamed liver cancers could be further subdivided based on their immunosuppression mechanisms. Each subclass had unique associations with key clinicopathological features of liver cancer.
3.4. Somatic alterations in the immunosuppression subclasses
Somatic genome alterations in tumor cells have opposing effects on tumor immunity and TME. Somatic mutations generate neoantigens and elicit attack by cytotoxic T cells, while somatic alterations of immune-modulating genes promote immune evasion by tumor cells [37], [38], [39]. To determine the associations between somatic alterations and the immunosuppression subclasses, we compared the somatic alterations detected by WGS with the immune signatures and subclasses of liver cancer.
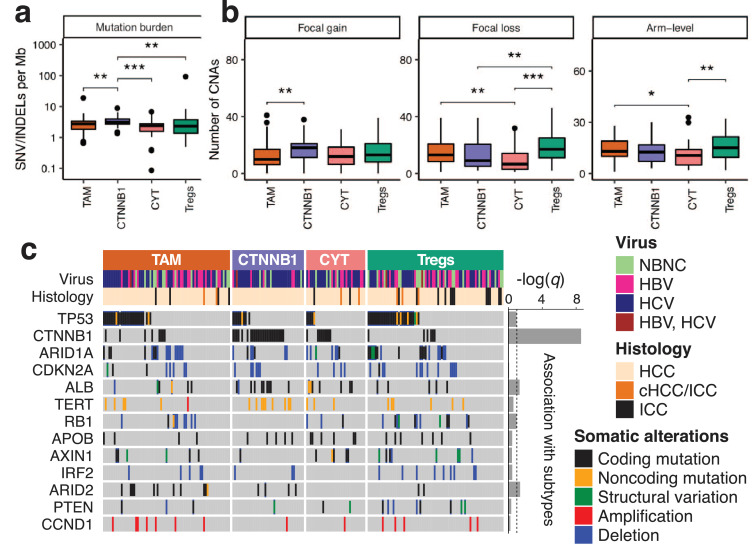
We first examined the mutation burden, which is the number of somatic single nucleotide variants (SNVs) and insertions/deletions (INDELs) per megabase (Mb) of coding regions. Previous studies found that highly mutated tumors are immunologically active in lung and colorectal cancers, among others [24,40]. However, in our liver cancer data, the non-inflamed CTNNB1 subclass had a higher mutation burden than the other subclasses (median 3.09 and 2.41 mutations per Mb, respectively; p = 0.0004) (Fig. 4a).
Fig. 4.
Somatic alterations in the immunosuppression subclasses. (a, b) The number of somatic alterations in the immunosuppression subclasses. (a) Mutation burdens and (b) somatic copy number alterations. Mutation burdens were computed for coding regions and included silent mutations. Non-significant (p ≥ 0.05) pairwise comparisons are omitted for clarity. ***p < 0.001; **p < 0.01; *p < 0.05. (c) Driver mutations and the immunosuppression subclasses of liver cancer. ‒log(q) is a measure of association between the mutations and the subclasses, where q denotes the adjusted p-value computed using Fisher's exact test and the Benjamini–Hochberg method. Dotted line shows q = 0.1.
We also looked at the total number of somatic copy number alterations (CNAs). A previous pan-cancer study reported a negative correlation between tumor aneuploidy and immune cell infiltration [41]. Consistent with this report, the lowest levels of arm-level CNAs were found in the CYT subclass in our liver cancer data (p = 0.0054) (Fig. 4b). However, the highest levels of arm-level CNAs were obtained in the Treg subclass (p = 0.014), which is another inflamed subclass in our classification. The results of the mutation burden and CNA analysis indicated the uniqueness of inflammation in liver cancer compared to other types of cancer.
Next, we investigated the somatic alterations of individual genes (Fig. 4c). Among 13 driver genes of liver cancers, the mutations of CTNNB1, ALB, and ARID2 were found to have a statistically significant association with the immunosuppressive subclasses (q < 0.05). The mutations of CTNNB1 and ALB were overrepresented in the CTNNB1 subclass (OR 11.2 and 2.98; p = 3.4 × 10−10 and p = 0.017, respectively). The mutation of ARID2 was overrepresented in the TAM subclass (OR 4.34; p = 0.0054). The ALB gene encodes the albumin protein, which appears to promote an inflammatory response by sequestering immunosuppressive prostaglandin E2 [42].
The ARID2 gene encodes a subunit of the chromatin remodeling complex PBAF, which is mutated in various cancers. In our liver cancer data, somatic alterations of ARID2 were found in 17 cases and associated with reduced levels of CYT (Supplementary Fig. 7a). GSEA showed that ARID2-mutated tumors had weaker inflammatory and interferon-γ responses than ARID2 wild-type tumors (FDR < 0.001) (Supplementary Fig. 7b). In contrast, the estimated fraction of macrophages M2 was significantly increased in ARID2-mutated tumors (Supplementary Fig. 7c and d). These results confirmed the tripartite association between low inflammation, TAM, and mutation of ARID2 in liver cancer.
3.5. Knockout of ARID2 impaired chemokine production in HCC cell lines
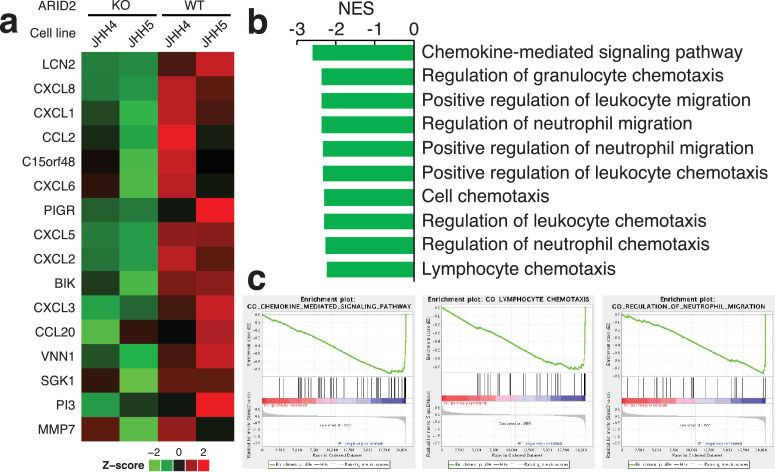
To investigate the immunological role of ARID2 in liver cancer, we analyzed the gene expression profiles of ARID2-knockout HCC cell lines. JHH4 and JHH5 are two human HCC-derived cell lines that express wild-type ARID2 genes, according to the Cancer Cell Line Encyclopedia database. In a previous study, we constructed a ARID2 knockout of JHH4 and JHH5, and confirmed the loss of ARID2 protein expression [23]. The gene expression profiles of ARID2-knockout and ARID2 wild-type cells were compared using microarray. The gene expression levels of 16 genes was found to decrease by over eight-fold in the knockout cells compared to the wild-type cells (Fig. 5a), while no gene expression increased by eight-fold. The 16 genes whose expression was found to decrease included 8 chemokines (CCL2, CCL20, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL8). Consistently, the gene sets related to the chemotaxis of lymphocytes and neutrophils was significantly downregulated in the ARID2-knockout cells (FDR < 0.001) (Fig. 5b and c). To validate this microarray result, we performed RNA-Seq on the JHH4 cells (Supplementary Fig. 8). Reduced expression levels were observed for 9 of 16 genes in the ARID2-knockout cells, where 5 out of these 9 genes showed chemotactic activity for neutrophils. These results suggested that ARID2 regulates chemokine production in liver cancer cells. Impaired chemokine signaling may account for the reduced inflammation in ARID2-mutated tumors.
Fig. 5.
Knockout of ARID2 in HCC cell lines reduced mRNA expression of cytokines. (a) mRNA expression profiles of ARID2-knockout and wild-type cell lines. Two HCC cell lines (JHH4 and JHH5) were analyzed. Only genes that showed changes over eight-fold are shown. (b, c) Gene set enrichment analysis comparing the ARID2-knockout and wild-type cells. (b) The ten most downregulated gene sets in the ARID2-knockout cells (FDR < 0.001). The gene sets in the Gene Ontology (GO) biological process were tested. No gene sets of GO were upregulated with FDR < 0.05. NES, normalized enrichment score. (c) Gene set enrichment plots for downregulated genes in the ARID2-knockout cells.
4. Discussion
Although most liver cancers develop from chronic hepatitis, their TME is immunosuppressive [43]. Various mechanisms have been proposed to try to explain the immunosuppressive environment of liver cancers. The infiltration of immunosuppressive leukocytes, such as Tregs, TAMs, and myeloid-derived suppressor cells, has been proposed as a possible mechanism for this TME. Cancer-associated fibroblasts are known to results in the dysfunction of T cells and NK cells in HCC [44,45], while the activation of WNT/β-catenin signaling mediates T cell exclusion [9,11]. Liver cancer cells also produce immunosuppressive molecules, such as indoleamine 2,3-dioxygenase (IDO), TGF-β, and IL-10. However, the distribution of and correlation between these mechanisms in clinical samples is unclear.
In this study, we analyzed the immunological features of 234 liver cancers using their gene expression profiles. We found that three well-established mechanisms for immunosuppression were mutually exclusive, rather than function in a cooperative manner. This finding enabled us to classify liver cancer into four immunosuppression subclasses. The TAM, CTNNB1, and Treg subclasses were named after their predominant immunosuppression mechanisms, whereas the CYT subclass may be deficient in immunosuppression. Among the four subclasses, the TAM and CTNNB1 subclasses had non-inflamed TME. Because they are infiltrated by low levels of T cells, these patients may not be promising subjects for immune checkpoint inhibition. A recent study reported that HCC with activating mutations of Wnt/β-catenin signaling responded poorly to immune checkpoint inhibitors [46]. The CTNNB1 subclass has already been described by previous studies, whereas the TAM subclass have not. Since the TAM subclass is associated with a poor prognosis, this subclass poses a therapeutic challenge and may benefit from immune therapies that target TAMs [47].
On the other hand, the CYT and Treg subclasses had inflamed TME. Previously, a reduction in intratumoral Tregs has been found to enhance the efficacy of immune checkpoint inhibitors in mouse models of skin, breast, and colon cancer [48], [49], [50]. Therefore, the CYT subclass may respond better to immune checkpoint inhibition than the Treg subclass.
Inflammation has complicated effects on the prognosis of liver cancer. Previous studies have reported that while inflammation in tumor confers a better survival [7], [8], [9], [10], [11], inflammation in non-tumorous livers is associated with late recurrence and reduced overall survival [9,51]. Consistent with these reports, we observed that inflammation in tumor and non-tumorous livers was associated with a better overall survival and a poor disease-free survival, respectively (Fig. 1). Since immune checkpoint inhibitors also activate immunity in hepatitis liver, their use in the adjuvant setting may require the monitoring of the long-term effects.
Our experiments suggested that ARID2 regulates chemokine production in HCC cells. ARID2 is a subunit of the chromatin remodeling complex PBAF. PBAF is a member of the SWI/SNF family of chromatin remodeling complexes, tasked with moving nucleosomes and regulating gene expression [52]. Previous studies have found that PBAF also regulates the transcription of immune-related genes. For example, ARID2 is required for interferon-responsive gene expression [53]. PBAF also downregulates immunosuppressive interleukin IL10 [54]. SWI/SNF complexes often regulate transcription through lineage specific enhancers [55]. ARID2 mutations in liver cancer may alter the chromatin structure of liver-specific enhancers and reduce the levels of chemokine expression.
In conclusion, this study demonstrated the diversity of immunosuppressive mechanisms in clinical specimens of liver cancers. The correlation between the tumor subclasses and immune suppression may facilitate the development of precision immune therapy for patients with liver cancer.
Funding
This work was partly supported by the Japan Agency for Medical Research and Development (AMED) Project for Cancer Research and Therapeutic Evolution (P-CREATE) (to H.N.). The funder had no role in study design, data collection, data analysis, interpretation, or writing of the report.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
The super-computing resource ‘SHIROKANE’ was provided by the Human Genome Center, The University of Tokyo (http://supcom.hgc.jp/). We would also like to acknowledge Keith A. Boroevich, Aya Sasaki, and technical staff in RIKEN for their technical supports and The PCAWG Tech working group for their mutational analysis.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102659.
Contributor Information
Masashi Fujita, Email: m-fujita@riken.jp.
Rui Yamaguchi, Email: ruiy@ims.u-tokyo.ac.jp.
Takanori Hasegawa, Email: t-hasegw@ims.u-tokyo.ac.jp.
Shu Shimada, Email: shimada.monc@tmd.ac.jp.
Koji Arihiro, Email: arihiro@hiroshima-u.ac.jp.
Shuto Hayashi, Email: s-haya@hgc.jp.
Kazuhiro Maejima, Email: kazuhiro.maejima@riken.jp.
Kaoru Nakano, Email: kaoru.nakano@riken.jp.
Akihiro Fujimoto, Email: afujimoto@m.u-tokyo.ac.jp.
Atsushi Ono, Email: atsushi-o@hiroshima-u.ac.jp.
Hiroshi Aikata, Email: aikata@hiroshima-u.ac.jp.
Masaki Ueno, Email: ma@wakayama-med.ac.jp.
Shinya Hayami, Email: shin-8@wakayama-med.ac.jp.
Hiroko Tanaka, Email: hiroko@hgc.jp.
Satoru Miyano, Email: miyano@ims.u-tokyo.ac.jp.
Hiroki Yamaue, Email: yamaue-h@wakayama-med.ac.jp.
Kazuaki Chayama, Email: chayama@hiroshima-u.ac.jp.
Kazuhiro Kakimi, Email: kakimi@m.u-tokyo.ac.jp.
Shinji Tanaka, Email: tanaka.monc@tmd.ac.jp.
Seiya Imoto, Email: imoto@ims.u-tokyo.ac.jp.
Hidewaki Nakagawa, Email: hidewaki@riken.jp.
Appendix. Supplementary materials
References
- 1.Bremnes R.M., Busund L-T, Kilvær T.L., Andersen S., Richardsen E., Paulsen E.E. The role of tumor-infiltrating lymphocytes in development, progression, and prognosis of non–small cell lung cancer. J Thorac Oncol. 2016;11:789–800. doi: 10.1016/j.jtho.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Mlecnik B., Tosolini M., Kirilovsky A., Berger A., Bindea G., Meatchi T. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29:610–618. doi: 10.1200/JCO.2010.30.5425. [DOI] [PubMed] [Google Scholar]
- 3.Goode E.L., Block M.S., Kalli K.R., Vierkant R.A., Chen W., Fogarty Z.C. Dose-response association of CD8 + tumor-infiltrating lymphocytes and survival time in high-grade serous ovarian cancer. JAMA Oncol. 2017;3 doi: 10.1001/jamaoncol.2017.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fridman W.H., Pagès F., Saut̀s-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 5.Cristescu R., Mogg R., Ayers M., Albright A., Murphy E., Yearley J. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018:362. doi: 10.1126/science.aar3593. (80-)eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ally A., Balasundaram M., Carlsen R., Chuah E., Clarke A., Dhalla N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341. doi: 10.1016/j.cell.2017.05.046. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chew V., Tow C., Teo M., Wong H.L., Chan J., Gehring A. Inflammatory tumour microenvironment is associated with superior survival in hepatocellular carcinoma patients. J Hepatol. 2010;52:370–379. doi: 10.1016/j.jhep.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Chew V., Chen J., Lee D., Loh E., Lee J., Lim K.H. Chemokine-driven lymphocyte infiltration: an early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut. 2012;61:427–438. doi: 10.1136/gutjnl-2011-300509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sia D., Jiao Y., Martinez-Quetglas I., Kuchuk O., Villacorta-Martin C., Castro de Moura M. Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology. 2017;153:812–826. doi: 10.1053/j.gastro.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Gabrielson A., Wu Y., Wang H., Jiang J., Kallakury B., Gatalica Z. Intratumoral CD3 and CD8 T-cell densities associated with relapse-free survival in hcc. Cancer Immunol Res. 2016;4:419–430. doi: 10.1158/2326-6066.CIR-15-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurebayashi Y., Ojima H., Tsujikawa H., Kubota N., Maehara J., Abe Y. Landscape of immune microenvironment in hepatocellular carcinoma and its additional impact on histological and molecular classification. Hepatology. 2018;68:1025–1041. doi: 10.1002/hep.29904. [DOI] [PubMed] [Google Scholar]
- 12.El-Khoueiry A.B., Sangro B., Yau T., Crocenzi T.S., Kudo M., Hsu C. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu A.X., Finn R.S., Edeline J., Cattan S., Ogasawara S., Palmer D. Pembrolizumab in patients with advanced hepatocellular carcinoma previously treated with sorafenib (KEYNOTE-224): a non-randomised, open-label phase 2 trial. Lancet Oncol. 2018;2045:1–13. doi: 10.1016/S1470-2045(18)30351-6. [DOI] [PubMed] [Google Scholar]
- 14.Zheng C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X. Landscape of infiltrating t cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356. doi: 10.1016/j.cell.2017.05.035. .e16. [DOI] [PubMed] [Google Scholar]
- 15.Gao Q., Qiu S.J., Fan J., Zhou J., Wang X.Y., Xiao Y.S. Intratumoral balance of regulatory and cytotoxic t cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25:2586–2593. doi: 10.1200/JCO.2006.09.4565. [DOI] [PubMed] [Google Scholar]
- 16.Ding T., Xu J., Wang F., Shi M., Zhang Y., Li S.-.P. High tumor-infiltrating macrophage density predicts poor prognosis in patients with primary hepatocellular carcinoma after resection. Hum Pathol. 2009;40:381–389. doi: 10.1016/j.humpath.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Fujimoto A., Furuta M., Totoki Y., Tsunoda T., Kato M., Shiraishi Y. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 18.Yung CK, O'Connor BD, Yakneen S, Zhang J, Ellrott K, Kleinheinz K, et al. Large-scale uniform analysis of cancer whole genomes in multiple computing environments. BioRxiv2017:161638. doi: 10.1101/161638. [DOI]
- 19.Fonseca NA, Petryszak R, Marioni J, Brazma A. iRAP – an integrated RNA-seq analysis pipeline. BioRxiv2014:005991. doi: 10.1101/005991. [DOI]
- 20.Bindea G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Chiang D.Y., Villanueva A., Hoshida Y., Peix J., Newell P., Minguez B. Focal gains of vegfa and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshida Y., Nijman S.M.B., Kobayashi M., Chan J.A., Brunet J.P., Chiang D.Y. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oba A., Shimada S., Akiyama Y., Nishikawaji T., Mogushi K., Ito H. ARID2 modulates dna damage response in human hepatocellular carcinoma cells. J Hepatol. 2017;66:942–951. doi: 10.1016/j.jhep.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013:4. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furuta M., Ueno M., Fujimoto A., Hayami S., Yasukawa S., Kojima F. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J Hepatol. 2017;66:363–373. doi: 10.1016/j.jhep.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 27.Goh G.B.B., Chang P.E., Tan C.K. Changing epidemiology of hepatocellular carcinoma in Asia. Best Pract Res Clin Gastroenterol. 2015;29:919–928. doi: 10.1016/j.bpg.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Murakami J., Shimizu Y., Kashii Y., Kato T., Minemura M., Okada K. Functional B-cell response in intrahepatic lymphoid follicles in chronic hepatitis c. Hepatology. 1999;30:143–150. doi: 10.1002/hep.510300107. [DOI] [PubMed] [Google Scholar]
- 29.Spranger S., Bao R., Gajewski T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 2015;523:231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 30.Calderaro J., Couchy G., Imbeaud S., Amaddeo G., Letouzé E., Blanc J.F. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol. 2017;67:727–738. doi: 10.1016/j.jhep.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Fu J., Xu D., Liu Z., Shi M., Zhao P., Fu B. Increased regulatory t cells correlate with CD8 T-cell impairment and poor survival in hepatocellular carcinoma patients. Gastroenterology. 2007;132:2328–2339. doi: 10.1053/j.gastro.2007.03.102. [DOI] [PubMed] [Google Scholar]
- 32.Chew V., Lai L., Pan L., Lim C.J., Li J., Ong R. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci. 2017;114:E5900–E5909. doi: 10.1073/pnas.1706559114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathai A.M., Kapadia M.J., Alexander J., Kernochan L.E., Swanson P.E., Yeh M.M. Role of foxp3-positive tumor-infiltrating lymphocytes in the histologic features and clinical outcomes of hepatocellular carcinoma. Am J Surg Pathol. 2012;36:980–986. doi: 10.1097/PAS.0b013e31824e9b7c. [DOI] [PubMed] [Google Scholar]
- 34.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noy R., Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coulouarn C., Factor V.M., Thorgeirsson S.S. Transforming growth factor-β gene expression signature in mouse hepatocytes predicts clinical outcome in human cancer. Hepatology. 2008;47:2059–2067. doi: 10.1002/hep.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kataoka K., Shiraishi Y., Takeda Y., Sakata S., Matsumoto M., Nagano S. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature. 2016;534:402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 38.Gao J., Shi L.Z., Zhao H., Chen J., Xiong L., He Q. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-ctla-4 therapy. Cell. 2016;167:397–404.e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sade-Feldman M., Jiao Y.J., Chen J.H., Rooney M.S., Barzily-Rokni M., Eliane J.P. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun. 2017:8. doi: 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grasso C.S., Giannakis M., Wells D.K., Hamada T., Mu X.J., Quist M. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov. 2018;8:730–749. doi: 10.1158/2159-8290.CD-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davoli T., Uno H., Wooten E.C., Elledge S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science. 2017:355. doi: 10.1126/science.aaf8399. (80-) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Brien A.J., Fullerton J.N., Massey K.A., Auld G., Sewell G., James S. Immunosuppression in acutely decompensated cirrhosis is mediated by prostaglandin E2. Nat Med. 2014;20:518–523. doi: 10.1038/nm.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prieto J., Melero I., Sangro B. Immunological landscape and immunotherapy of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2015;12:681–700. doi: 10.1038/nrgastro.2015.173. [DOI] [PubMed] [Google Scholar]
- 44.Li T., Yang Y., Hua X., Wang G., Liu W., Jia C. Hepatocellular carcinoma-associated fibroblasts trigger nk cell dysfunction via PGE2 and IDO. Cancer Lett. 2012;318:154–161. doi: 10.1016/j.canlet.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 45.Cheng Y., Li H., Deng Y., Tai Y., Zeng K., Zhang Y. Cancer-associated fibroblasts induce PDL1+ neutrophils through the IL6-STAT3 pathway that foster immune suppression in hepatocellular carcinoma. Cell Death Dis. 2018:9. doi: 10.1038/s41419-018-0458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M, et al. Prospective genotyping of hepatocellular carcinoma: clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res2018:1–12. doi: 10.1158/1078-0432.CCR-18-2293. [DOI] [PMC free article] [PubMed]
- 47.Poh A.R., Ernst M. Targeting macrophages in cancer: from bench to bedside. Front Oncol. 2018;8:1–16. doi: 10.3389/fonc.2018.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simpson T.R., Li F., Montalvo-Ortiz W., Sepulveda M.A., Bergerhoff K., Arce F. Fc-dependent depletion of tumor-infiltrating regulatory t cells co-defines the efficacy of anti–CTLA-4 therapy against melanoma. J Exp Med. 2013;210:1695–1710. doi: 10.1084/jem.20130579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selby M.J., Engelhardt J.J., Quigley M., Henning K.A., Chen T., Srinivasan M. Anti-CTLA-4 antibodies of igg2a isotype enhance antitumor activity through reduction of intratumoral regulatory t cells. Cancer Immunol Res. 2013;1:32–42. doi: 10.1158/2326-6066.CIR-13-0013. [DOI] [PubMed] [Google Scholar]
- 50.Taylor N.A., Vick S.C., Iglesia M.D., Brickey W.J., Midkiff B.R., McKinnon K.P. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J Clin Investig. 2017;127:3472–3483. doi: 10.1172/JCI90499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu J.C., Huang Y.H., Chau G.Y., Su C.W., Lai C.R., Lee P.C. Risk factors for early and late recurrence in hepatitis B-related hepatocellular carcinoma. J Hepatol. 2009;51:890–897. doi: 10.1016/j.jhep.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Mathur R., Roberts C.W.M. SWI/SNF (BAF) complexes: guardians of the epigenome. Annu Rev Cancer Biol. 2018;2:413–427. [Google Scholar]
- 53.Yan Z., Cui K., Murray D.M., Ling C., Xue Y., Gerstein A. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wurster A.L., Precht P., Becker K.G., Wood W.H., Zhang Y., Wang Z. IL-10 transcription is negatively regulated by BAF180, a component of the swi/snf chromatin remodeling enzyme. BMC Immunol. 2012;13:9. doi: 10.1186/1471-2172-13-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alver B.H., Kim K.H., Lu P., Wang X., Manchester H.E., Wang W. The swi/snf chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun. 2017;8:14648. doi: 10.1038/ncomms14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS and RNA-Seq reads for matched tumor and non-tumor tissues are available in the European Genome-phenome Archive database (accession number EGAS00001000678). The gene expression data for cell lines were deposited to the NCBI's Gene Expression Omnibus. The accession number is GSE144021 for microarray and GSE143941 for RNA-Seq.