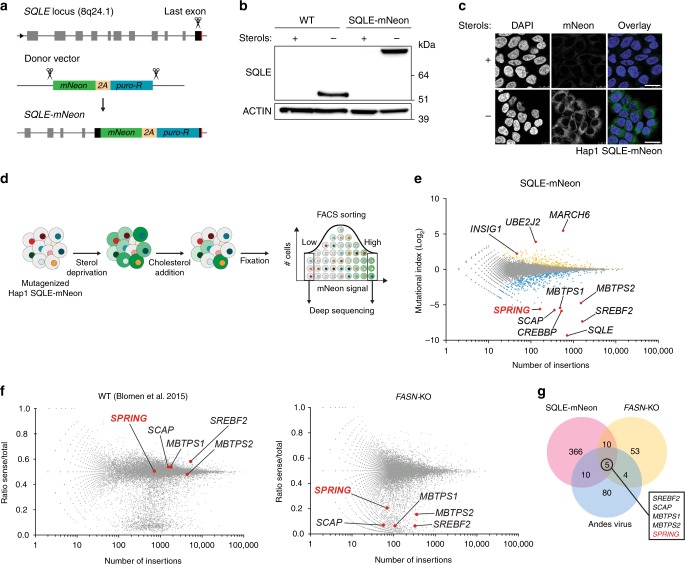
Fig. 1. Haploid mammalian genetic screens identify SPRING as an SREBP regulator.
a Schematic illustration of CRISPR/Cas9-mediated targeting of the endogenous SQLE locus for in-frame integration of mNeon-2A-PURO. b Hap1 control and Hap1 SQLE-mNeon cells were cultured in the presence or absence of sterols and total cell lysates were immunoblotted as indicated. c Hap1 SQLE-mNeon cells were grown as in b on coverslips and subsequently fixed and counterstained with DAPI (nucleus). Cells were imaged and representative images are shown; scale bar, 25 μm. d Schematic depiction of SQLE-mNeon screen: a library of mutant Hap1 SQLE-mNeon cells was cultured in sterol-depletion medium for 24 h to induce SQLE expression. During the last 6 h β-methyl-cyclodextrin-cholesterol was added to initiate sterol-stimulated degradation of SQLE after which mNeonHIGH and mNeonLOW cells were isolated by FACS. e The log mutational index-scores (see Methods section) were plotted against the number of trapped alleles per gene. Statistically significant hits (p < 0.05) in the isolated mNeonHIGH and mNeonLOW cell populations are indicated in yellow and blue, respectively. f Comparison of gene-essentiality screens between control Hap1 cells and Hap1-FASNKO cells. Per gene, the ratio of sense/total orientation gene-trap insertions (y-axis) and the total number of insertions in a particular gene (x-axis) are plotted. FASN synthetic lethality screens show that SCAP, SREBF2, MBTPS1/2, and SPRING are significantly depleted in FASNKO cells. g Venn diagram indicating the number of unique and commonly identified genes in 3 individual SREBP-focused screens. b, c Representative images of three independent experiments are shown.