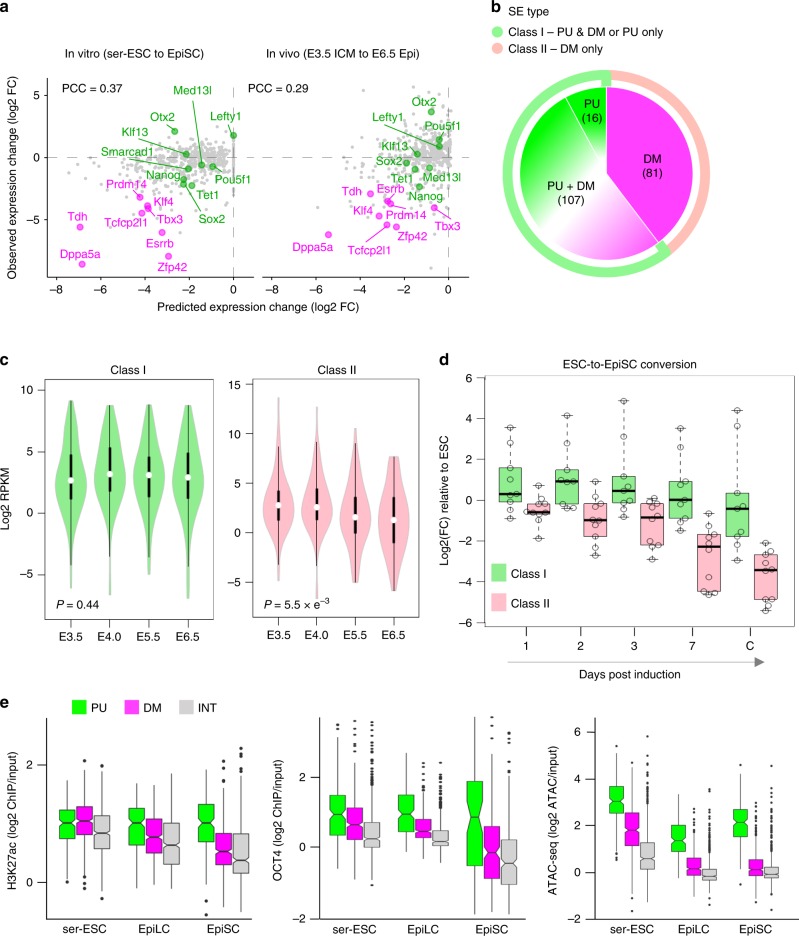
Fig. 3. Partitioning of enhancer units within SEs as constitutively active or decommissioned in primed cells.
a Predicted and observed gene expression fold change (log2) of SE target genes in vitro (left) and in vivo (right). The predicted fold change is the negative product of the interaction intensity (log2 normalised reads) in ESCs (serum/LIF; ser) and CpG methylation changes (percent change) at PU and DM subregions from ESCs to EpiSCs (in vitro) or E3.5 ICM to E6.5 Epi (in vivo; see Methods section and Supplementary Data 1). Well known pluripotency genes associated with different categories of SE subregions are highlighted in magenta (DM) and in green (PU). PCC, Pearson’s correlation coefficient. b Venn diagram depicting SE subregion composition and number of SEs falling into class I (light green) and class II (pink) categories. Class I SEs contain PU or PU + DM subregions, while class II SEs only contain DM subregions. c Expression (log2 RPKM) of all closest interacting gene promoters associated with class I or class II SEs in epiblast cells in vivo at different developmental times (from E3.5 to E6.5)22. p-Values (Kruskal–Wallis test) are indicated. d Expression changes (log2(fold change)) relative to ESCs (serum/LIF) of selected genes associated with class I (light green) or class II (pink) SEs during the conversion of ESCs into EpiSCs in vitro (RT-qPCR). Class I candidates are: Lefty1, Pou5f1, Otx2, Smarcad1, Sox2, Tet1, Klf13, Med13l and Nanog; class II candidates are: Esrrb, Klf4, Prdm14, Tbx3, Tdh, Tet2, Tfcp2l1, Klf2, Klf5 and Zfp42. Statistical analysis was performed using non parametric tests for repeated measures data in factorial designs. The factorial design chosen corresponds to f1.ld.f1 in nparLD R-package. Kinetics of the two classes were statistically different (p = 4.5 × 10−12, Wald test). Data represent three independent conversion experiments (n = 3). Source data are provided as Supplementary Data 10. e H3K27ac deposition, OCT4 binding and chromatin accessibility (ATAC-seq) of PU, DM and INT subregions in ESCs (serum/LIF; ser), EpiLCs and EpiSCs. Publicly available datasets used are listed in Supplementary Data 1.