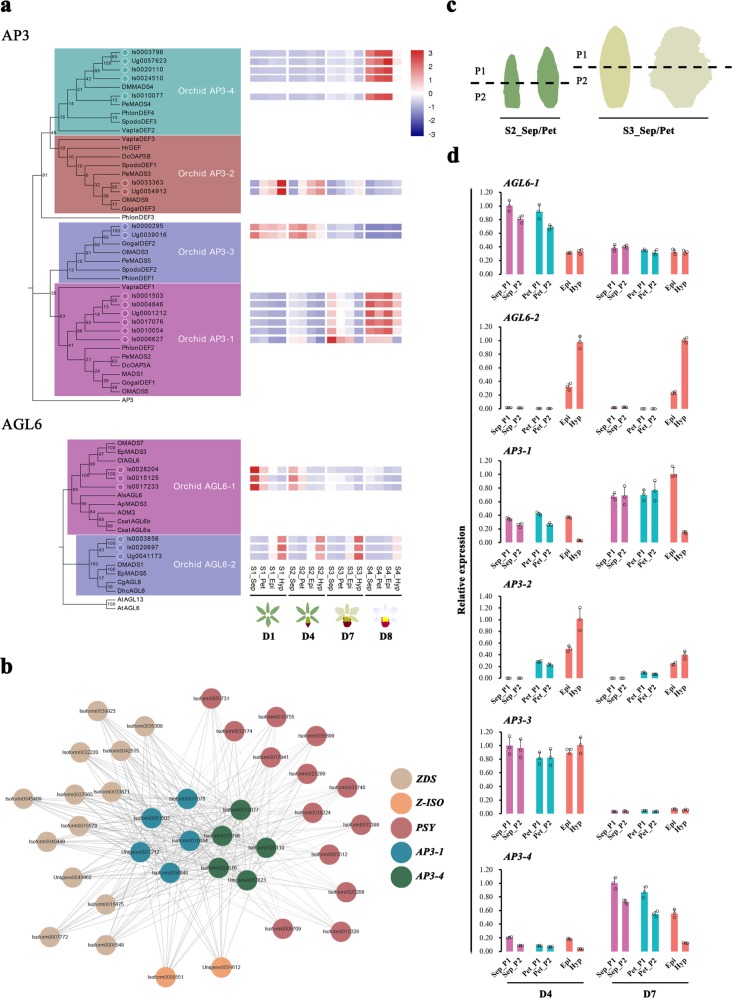
Fig. 5. Spatiotemporal expression analysis of the AP3- and AGL6-like genes in the perianth and lip segments during floral development.
a Phylogenetic trees based on the neighbour-joining method were inferred from the amino acid sequences of KOVA AP3- and AGL6-like MADS-box proteins and those of other plants (left). According to the FPKM values of these KOVA MADS-box genes, their expression profiles in flower segments during floral development were described via heatmap analysis (right). The FPKM value of each isoform/unigene is the mean of three biological replicates. b The brown module shows that AP3-1s and AP3-4s presented a co-expression relationship with some CBP structural genes during floral development according to WGCNA. c Sampling strategy for the sepals and petals at D4 (left) and D7 (right), which was similar to the sampling strategy for the lip. These samples were used for qRT-PCR analysis to explore the AP3- and AGL6-like MADS-box gene expression profiles. d The qRT-PCR results showed that the differences in the expression levels of the AP3- and AGL6-like genes between the perianth segments were smaller than those in the lip segments. The data are the mean ± SD from three biological replicates.