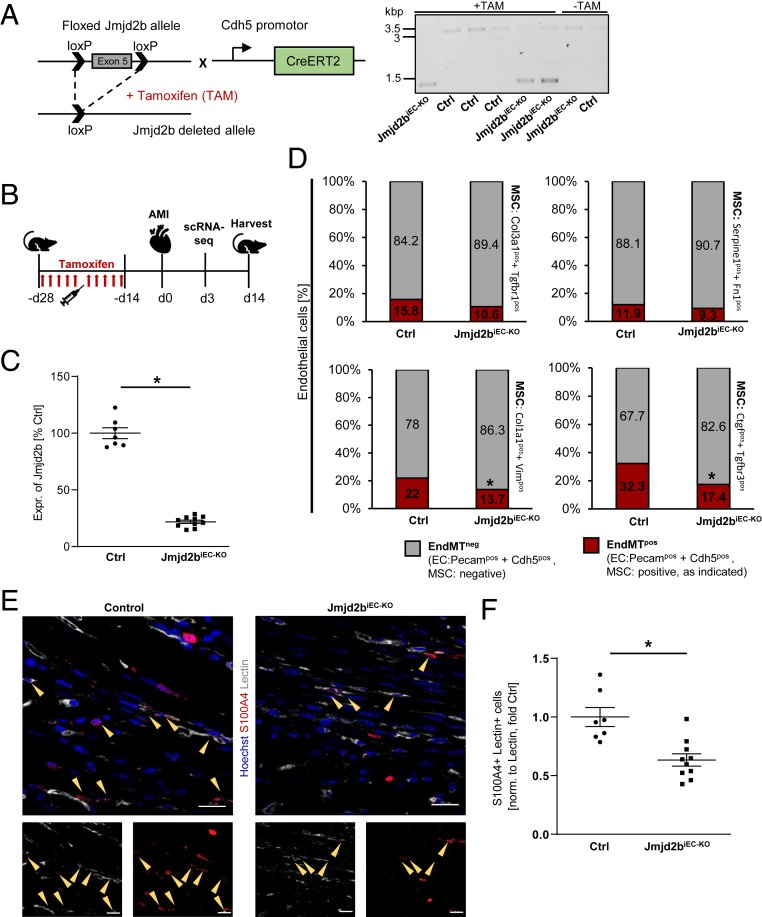
Fig. 3.
Jmjd2b is a positive regulator of EndMT in vivo. (A) Knockout strategy and DNA gel of the floxed Jmjd2b and the Cdh5-CreERT2 allele. Recombination after TAM treatment led to the deletion of exon 5 in the Jmjd2b ORF. Jmjd2b DNA in total heart of Ctrl (3.1 kbp) or Jmjd2biEC-KO (1.4 kbp) mice, with or w/o TAM. (B) Schema of TAM-induced EC deletion of Jmjd2b with a subsequent AMI induction. (C) Jmjd2b mRNA of isolated EC of control (Ctrl) or Jmjd2biEC-KO mice, determined by RT-qPCR, depicted as percentage of Ctrl, n = 7–10. (D) Single-cell RNA sequencing showing percentage of cells positive [≥1 unique molecular identifiers (UMI) counts] for EC marker (Pecam1 and Cdh5) only (gray) and EC plus mesenchymal cell marker (MSC) gene expression (red) 3 d post-AMI. (E) Representative images of IHC stainings of Ctrl and Jmjd2biEC-KO mice hearts 14 d post-AMI. Hoechst appears in blue, lectin in white, and S100A4 in red, scale bar = 20 μM. (F) S100A4+ and lectin+ cells were counted in the sections of Ctrl or Jmjd2biEC-KO mice, normalized to area of lectin, depicted as fold Ctrl, n = 7–10. Data are depicted ±SEM. Student’s t test (C and F), χ2 test with Yates correction (D); *P < 0.05.