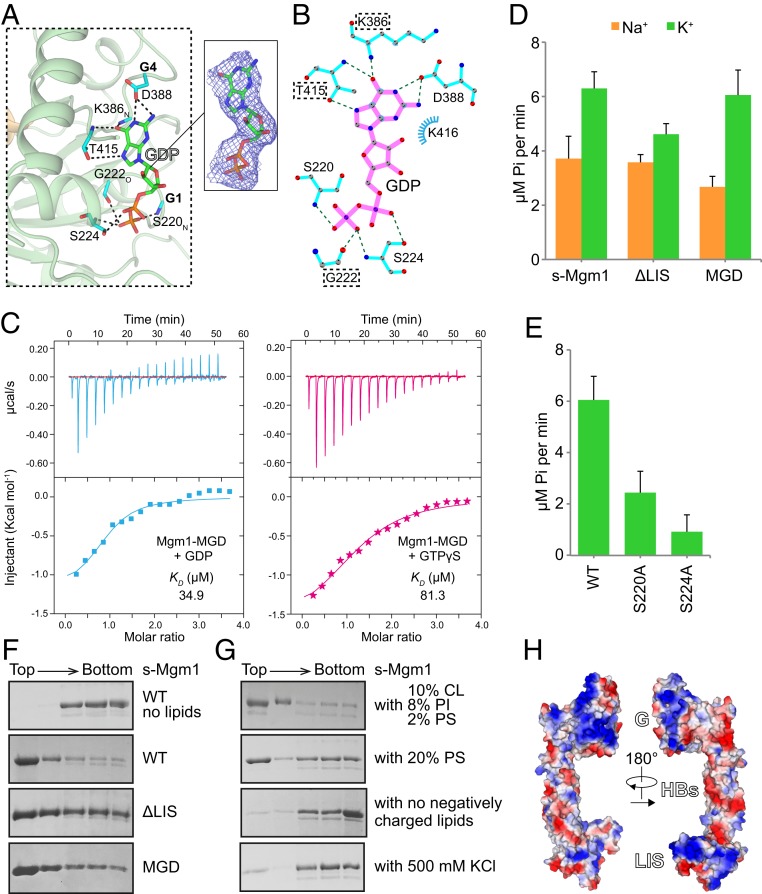
Fig. 2.
GTP hydrolysis and lipid interactions of Mgm1. (A) The active site of Mgm1 is shown in stick form with key residues highlighted. The 2Fo − Fc electron density maps (1.0σ contour) of GDP are shown as wire mesh (blue). (B) Interaction map of the catalytic core generated by LIGPLOT. (C) Binding affinity of GDP or GTPγS for Mgm1-MGD as measured by ITC. A 2 mM nucleotide solution was titrated stepwise into 0.1 mM protein. The dissociation constant (KD) is given in Inset. The data are representative of at least three repetitions. (D) The GTPase activity of various Mgm1 constructs was measured in the presence of 500 mM NaCl/KCl and 5 mM MgCl2. A total of 5 μM of protein was used for each sample. GTP hydrolysis was measured by phosphate release at saturating GTP concentrations (0.5 mM). Data are presented as the mean ± SD of nine measurements from three independent experiments. (E) As in D, but with WT or mutant s-Mgm1 in KCl-containing buffer. (F) Liposome flotation assay with various Mgm1 constructs. Liposomes (2 mM, POPC:POPE:CL:PI:DOPS:Rhodamine-PE = 55:23.5:10:8:2:1.5) were mixed with 2 μM purified protein at room temperature for 30 min. Fractions collected after centrifugation were analyzed by SDS/PAGE and Coomassie blue staining. The data are representative of at least three repetitions. (G) As in F, but with liposomes of different lipid compositions or different buffer conditions. (H) Electrostatic analysis of s-Mgm1. Negatively charged surfaces are colored in red and positively charged surfaces in blue. The s-Mgm1 molecule is viewed from both sides.