Abstract
Background
Local relapses and metastases are primary causes of death in lung cancer patients. In the present study, we aimed to develop a prognostic signature based on metastasis‐associated lncRNAs in patients with lung adenocarcinoma (LUAD).
Methods
Firstly, the potential metastasis‐associated lncRNAs were identified by analyzing high‐throughput data from The Cancer Genome Atlas (TCGA), and based on which, an lncRNA signature was constructed for prediction of relapse in LUAD patients using Cox proportional hazards regression analysis. Moreover, the prognostic performance of the lncRNA signature was evaluated using Kaplan‐Meier survival analysis, time‐dependent receiver operating characteristic (ROC) curve and Cox analysis, respectively. In addition, the potential metastasis‐associated function of these six lncRNAs was confirmed by lncRNA over‐expression or depletion and in vitro transwell assays in LUAD cells.
Results
An lncRNA signature consisting of six most important prognostic factors (LINC01819, ZNF649‐AS1, HNF4A‐AS1, FAM222A‐AS1, LINC02323 and LINC00672) was developed. The signature was an independent predictor for patients' relapse‐free survival (RFS), which could provide higher tumor relapse prediction capability compared with the TNM staging system at three years and five years, respectively (P = 0.0209 and P = 0.0468). Furthermore, the combination of this lncRNA signature and TNM stage had better prognostic value than TNM stage alone at three and five years, respectively (P = 0.0006 and P = 0.0096). Additionally, all the lncRNAs of the signature had a regulatory role in the LUAD cell mobility.
Conclusions
This novel six‐lncRNA signature had considerable prognostic value for prediction of relapse in LUAD patients.
Key points
Significant findings of the study
The unique metastasis‐associated lncRNA signature was related to tumor metastasis and prognosis in LUAD patients.
What this study adds
This signature had considerable prognostic value for prediction of relapse in LUAD patients.
Keywords: lncRNA signature, lung adenocarcinoma, metastasis, prediction, prognosis
Introduction
Lung cancer is the leading cause of cancer mortality worldwide.1 Lung adenocarcinoma (LUAD) is the most diagnosed subtype of lung cancer, which has different genomic alterations compared with other subtypes of lung cancer.2, 3 Identification of a subset of high‐risk patients for relapse may lead to more appropriate treatment and improved survival. However, current prognostication methods which largely rely on the tumor‐node‐metastasis (TNM) staging system are limited because the prognoses within the same TNM stage vary greatly. In order to help clinicians understand relapse risk in various patient subgroups, development of reliable markers for predicting the relapse risk in patients is urgently required.
As a large class of RNA molecules of more than 200 nucleotides in length, long non‐coding RNAs (lncRNAs) do not encode proteins, which may exert their functions by specifically binding to either RNA/DNA or proteins.4 Increasing evidence shows that lncRNAs play a critical role in cancer biology. Dysregulation of lncRNAs often serves as an independent predictor for poor prognosis of cancer patients.5, 6, 7, 8, 9 Our group has previously reported that circulating lncRNAs act as potential novel biomarkers for diagnosis and prognosis of non‐small cell lung cancer.10 Several recent studies have focused on the involvement of lncRNAs in tumor metastasis because these lncRNAs have shown great promise in the discovery of new biomarkers for predicting prognosis in individual patients.11, 12, 13 Some lncRNAs, such as HCP5, CAR10, NEAT1 and MALAT1, promote lung cancer metastasis via various mechanisms.7, 14, 15 These findings suggest that the tumor metastasis‐associated lncRNAs in lung cancer have potential prognostic implications. In the present study, we hypothesized that there were metastasis‐associated lncRNAs in LUAD which were differentially expressed between metastatic and nonmetastatic primary cancer tissues, and the dysregulated lncRNA expression regulated the aggressiveness, or in general, the prognosis. However, there is no comprehensive analysis of predictive biomarkers for prognosis based on metastasis‐associated lncRNA expression profiles in LUAD patients.
Our group and others have reported that the combination of a panel of multiple biomarkers, rather than just a single factor, may yield more powerful information in the clinical setting.16, 17, 18, 19 In the current study, we employed a large cohort of LUAD patients from The Cancer Genome Atlas (TCGA) project and identified a novel metastasis‐ associated lncRNA signature for predicting relapse‐free survival (RFS) in LUAD patients. Additionally, the potential role of these LncRNAs in the metastasis of lung cancer was determined using migration and invasion in vitro assays.
Methods
Patient information and data preprocessing
Data for selected samples of 347 LUAD patients with complete lncRNAs data, clinicopathological characteristics and follow‐up information (records of recurrence), were downloaded from the TCGA database (https://cancergenome.nih.gov/). The RNA‐seq dataset was annotated with the HUGO Gene Nomenclature Committee (HGNC, http://www.genenames.org/). The data of lncRNA expressions were processed by the background correction and normalization using edgeR in the Bioconductor package (version 3.6). A total of 3483 lncRNAs were obtained, which were matched to the clinicopathological information and follow‐up data (Table S1).
Construction of the prognostic risk formula based on metastasis‐associated lncRNAs
Patients were divided into two groups according to their metastatic status, including lymphatic and hematogenous metastasis. The differentially expressed lncRNAs between LUAD patients with (n = 158) and without metastasis (n = 189) were identified by EdgeR in R language of |logFC| > 1 and FDR < 0.05. The patients were randomly divided into a training set (n = 231) and a testing set (n = 116). According to the stage analysis, the data of some patients was not available and was automatically separated. The training set was applied to identify the potential lncRNA biomarkers in the following analysis. Univariate Cox proportional hazards regression analysis was used to identify the association between the differentially expressed lncRNAs and RFS by the R package “survival”. The differentially expressed lncRNAs with a P‐value less than 0.05 were selected as candidate variables. Subsequently, the candidate variables were assessed by multivariate Cox regression analysis to identify the lncRNA biomarkers of the predictive model. The prognostic lncRNA signature was constructed based on the following formula:
where k was the number of prognostic lncRNAs, Wi was the estimated regression coefficient of the lncRNA in the multivariate Cox regression analysis, and Ei represented the expression value of the lncRNA. The lncRNAs with Wi > 0 were defined as high‐risk signatures while those with Wi < 0 were defined as protective lncRNAs. Using the mean risk score as the cutoff point, LUAD patients were stratified into high and low‐risk groups.
Evaluation of prognostic performance for selected lncRNA signature
The prognostic performance of the signature was evaluated in the training, testing and entire cohorts. The difference in RFS between the high‐ and low‐risk groups was evaluated by Kaplan‐Meier survival curves, and the statistical significance was determined by the two‐sided log‐rank test using the R package “survival”. The prognostic performance of the prognostic lncRNA signature was assessed by the area under the curve (AUC) of the receiver operating characteristic (ROC) curve using the R package “survivalROC” based on time‐dependent ROC analysis.20 Moreover, the independent predictors which were strikingly contributed to patients' RFS were identified using the univariate and multivariate Cox regression analyses.
Cell culture, vector construction and transfection
Four LUAD cell lines (A549, SPC‐A‐1, H1975 and H1299) were purchased from Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were maintained in DMEM (Gibco, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA), 50 U/mL penicillin and 0.1 mg/mL streptomycin at 37°C in a humidified atmosphere containing 5% CO2. After the cells reached the logarithmic growth phase, subsequent experiments were conducted.
Full‐length sequences of ZNF649‐AS1, HNF4A‐AS1, LINC01819, FAM222A‐AS1, LINC02323 and LINC00672 were amplified using specific primers. The primer sequences were listed in Table S2. The amplified fragments were subcloned into NheI/KpnI and XhoI sites of pcDNA3.1(+) vectors (Invitrogen, USA). The recombinant vectors were confirmed by restriction enzyme digestion and DNA sequencing (Biosune Biotechnology, Shanghai, Co., Ltd.). Short interfering RNA (siRNA) sequences, which specifically target LINC02323 transcript, were directly synthesized (Oligobio, Beijing, China). Scrambled sequence was used as a negative control of siRNA (si‐NC). Cells were transiently transfected with plasmid vectors, siRNA or si‐NC using siTran1.0 (ORIGENE). The sequences of siRNA were as follows: si‐ Linc02323, 5′‐GCCGGGTTGTTCTTTCCTT‐3′. si‐NC, 5′‐TTCTCCGAACGTGTCACGT‐3′. The over‐expression and depletion efficiencies were determined by quantitative real‐time polymerase chain reaction (qRT‐PCR).
RNA extraction, cDNA synthesis and qRT‐PCR
Total RNA was extracted from cells using Trizol reagent (Invitrogen, USA) according to the manufacturer's instructions. Briefly, 2 μg RNA was reversely transcribed to cDNA using Super RT cDNA synthesis kit (Toyobo). qRT‐PCR was performed on the 7500 Fast Real‐time PCR System (Applied Biosystems) using the Real Master Mix. Unique cDNA primers of lncRNAs used for qRT‐PCR analysis were listed in Table S2, and GAPDH was selected as a housekeeping gene. Briefly, after an initial denaturation step at 95°C for 10 minutes, amplifications were carried out with 40 cycles at a melting temperature of 95°C for 15 seconds and an annealing temperature of 60°C for 60 seconds. Relative expressions of target genes were calculated using the 2‐△△Ct method.
Transwell invasion and migration assay
Transwell assay was conducted using 8.0 μm Transwell Permeable Supports (Corning, USA) according to the manufacturer's instructions. After 24 hours of cell transfection, 5 × 104 A549 or 2 × 105 SPC‐A‐1 cells suspended in 100 μL serum‐free medium were plated in the upper chamber precoated with or without Matrigel Matrix (BD Biosciences), and 600 μL culture medium containing 10% FBS was added into the lower chamber. After incubation at 37°C for 24 hours, the nonmigrating cells were mechanically removed from the upper part of the filter with a cotton swab. Cells on the bottom surface of the membrane were fixed with 4% paraformaldehyde for 15 minutes and then stained with 0.1% crystal violet solution. The migrated cells were counted using a digital microscope (Olympus IX81, Japan) for statistical analysis.
Statistical analysis
All statistical analyses were conducted with R version 3.4.3 and GraphPad Prism 7.0 software. A volcano plot was drawn using the “ggplot2” package of R software. The associations of continuous and categorical variables were analyzed by the Mann‐Whitney U test and sχ2 test, respectively. Differential expressions of lnRNAs between LUAD patients with and without metastasis were analyzed using the edgeR package of R software. Univariate and multivariate Cox analyses were carried out to screen independent prognostic factors of RFS. The Cox regression coefficients were used to establish a prognostic risk formula based on metastasis‐associated lncRNAs. For survival analyses, the Kaplan‐Meier method was used to plot survival curves that were compared by log‐rank tests. Time‐dependent ROC analysis was applied to assess the predictive accuracy of the lncRNA signature. The primary end point was relapse. A value of P < 0.05 was considered statistically significant.
Results
Clinical characteristics
Baseline clinical and pathological characteristics are shown in Table 1, which were similar between the training and testing cohorts (all P > 0.05).
Table 1.
Baseline clinical and pathological characteristics of study patients
| Training set | Testing set | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number | Low risk | High risk | P‐value | Number | Low risk | High risk | P‐value | * P‐value | |
| Age (years) | 0.896 | 0.133 | 0.435 | ||||||
| Mean (SD) | 63.94 (10.36) | 64.04 (9.67) | 63.86 (11.02) | 64.86 (10.23) | 63.30 (10.33) | 66.17 (10.05) | |||
| Gender | 0.807 | 0.376 | 0.224 | ||||||
| Male | 104 (45.0%) | 49 (43.8%) | 55 (46.2%) | 61 (52.6%) | 25 (47.2%) | 36 (57.1%) | |||
| Female | 127 (55.0%) | 63 (56.2%) | 64 (53.8%) | 55 (47.4%) | 28 (52.8%) | 27 (42.9%) | |||
| Smoking history | 0.891 | 0.898 | 0.196 | ||||||
| Never smoke | 39(16.9%) | 20(17.9%) | 19(16.0%) | 29(25.0%) | 14(26.4%) | 15 (23.8%) | |||
| Ever smoke | 119 (51.5%) | 58 (51.8%) | 61 (51.3%) | 53 (45.7%) | 23 (43.4%) | 30 (47.6%) | |||
| Current smoke | 73 (31.6%) | 34 (30.4%) | 39 (32.8%) | 34 (29.3%) | 16 (30.2%) | 18 (28.6%) | |||
| Stage | 0.331 | 0.155 | 0.121 | ||||||
| I | 115 (50.9%) | 59 (53.6%) | 56 (48.3%) | 49 (42.6%) | 28 (52.8%) | 21 (33.9%) | |||
| II | 53 (23.5%) | 26 (23.6%) | 27 (23.3%) | 37 (32.2%) | 16 (30.2%) | 21 (33.9%) | |||
| III | 41 (18.1%) | 15 (13.6%) | 26 (22.4%) | 25 (21.7%) | 8 (15.1%) | 17 (27.4%) | |||
| IV | 17 (7.5%) | 10 (9.1%) | 7 (6.0%) | 4 (3.5%) | 1 (1.9%) | 3 (4.8%) | |||
| M stage | 0.628 | 0.199 | 0.083 | ||||||
| M0 | 189 (83.6%) | 89 (82.4%) | 100 (84.7%) | 96 (83.5%) | 44 (83.0%) | 52 (83.9%) | |||
| M1 | 17 (7.5%) | 10 (9.3%) | 7 (5.9%) | 3 (2.6%) | 0 (0.0%) | 3 (4.8%) | |||
| MX | 20 (8.8%) | 9 (8.3%) | 11 (9.3%) | 16 (13.9) | 9 (17.0%) | 7 (11.3%) | |||
| N stage | 0.018 | 0.443 | 0.353 | ||||||
| N0 | 139 (60.2%) | 70 (62.5%) | 69 (58.0%) | 60 (52.2%) | 31 (58.5%) | 29 (46.8%) | |||
| N1+ | 86 (37.2%) | 36 (32.1%) | 50 (42.0%)) | 52 (45.2%) | 21 (39.6%) | 31 (50.0%) | |||
| NX | 6 (2.6%) | 6 (5.4%) | 0 (0.0%) | 3 (2.6%) | 1 (1.9%) | 2 (3.2%) | |||
| T stage | 0.576 | 0.646 | 0.300 | ||||||
| T1‐T2 | 204 (88.3%) | 98 (87.5%) | 106 (89.1%) | 97 (83.6%) | 46 (86.8%) | 51 (81.0%) | |||
| T3‐T4 | 26 (11.3%) | 13 (11.6%) | 13 (10.9%) | 17 (14.7%) | 6 (11.3%) | 11 (17.5%) | |||
| TX | 1 (0.4%) | 1 (0.9%) | 0 (0.0%) | 2 (1.7%) | 1 (1.9%) | 1 (1.6%) | |||
P, statistical differences between the training and validation sets.
Identification of potential metastasis‐associated lncRNAs
Based on the lncRNA expression data from TCGA dataset, we compared the lncRNA expression profiles between LUAD patients with and without metastasis. A total of 735 lncRNAs were identified according to the threshold of |logFC| > 1 and FDR < 0.05 (Fig 1). These significantly differentially expressed lncRNAs were considered as candidate prognostic biomarkers for LUAD patients.
Figure 1.
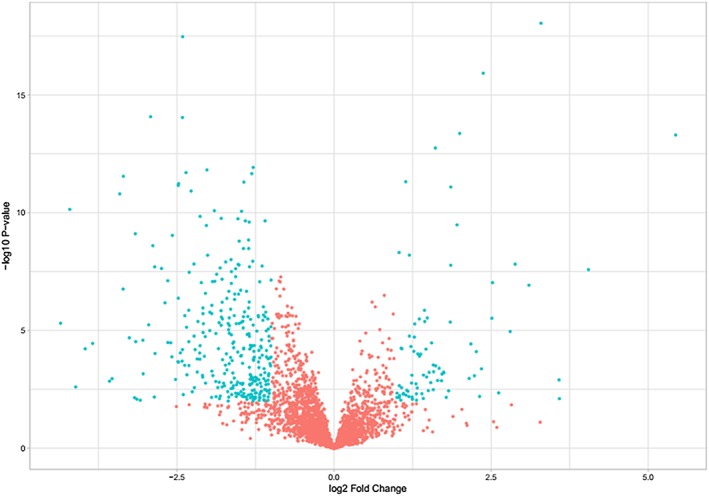
Volcano plot showing 735 differentially expressed lncRNAs between LUAD patients with and without metastasis from the TCGA cohort. Green color indicates differential lncRNAs, and red color represents undifferentiated lncRNAs.
Construction of a prognostic lncRNA signature
To determine the most important prognostic LncRNAs, 735 metastasis‐associated lncRNAs were initially subjected to univariate Cox proportional hazards regression analysis the training set consisting of 231 patients. A total of 43 lncRNAs were found to be significantly associated with the RFS of patients (P‐value < 0.05, Table S3) followed by the multivariate Cox regression analysis. Six lncRNAs (Table 2), including LINC01819, ZNF649‐AS1, HNF4A‐AS1, FAM222A‐AS1, LINC02323 and LINC00672, were subsequently selected to construct a signature, and there was no significant correlation between any pair of lncRNAs (Fig 2). Among these lncRNAs, three (LINC02323, ZNF649‐AS1 and HNF4A.AS1) with positive coefficient were risk factors owing to the close association between their high expression and shorter patients' RFS, whereas the remaining three (LINC01819, FAM222A‐AS1 and LINC00672) were protective factors. The expression level of each lncRNA in the signature was weighted by using the regression coefficients of multivariate Cox regression model, and the risk‐score formula for RFS prediction was developed as follows: RS = (0.191404 × the expression value of LINC02323) + (0.256783 × the expression value of ZNF649‐AS1) + (0.502202 × the expression value of HNF4A‐AS1) + (−0.319007 × the expression value of LINC01819) + (−0.306173 × the expression value of FAM222A‐AS1) + (−0.304856 × the expression value ofLINC00672).
Table 2.
Results of multivariate Cox regression analysis
| Name | Coefficient | HR | 95% CI | P‐value |
|---|---|---|---|---|
| LINC02323 | 0.191 | 1.211 | 1.024–1.433 | 0.026 |
| ZNF649.AS1 | 0.257 | 1.292 | 1.023–1.633 | 0.031 |
| HNF4A.AS1 | 0.502 | 1.652 | 1.052–2.595 | 0.029 |
| LINC01819 | −0.319 | 0.727 | 0.582–0.908 | 0.005 |
| FAM222A.AS1 | −0.306 | 0.736 | 0.625–0.868 | <0.001 |
| LINC00672 | −0.305 | 0.737 | 0.593–0.916 | 0.006 |
CL, confidence interval; HR, Hazard ratio.
Figure 2.
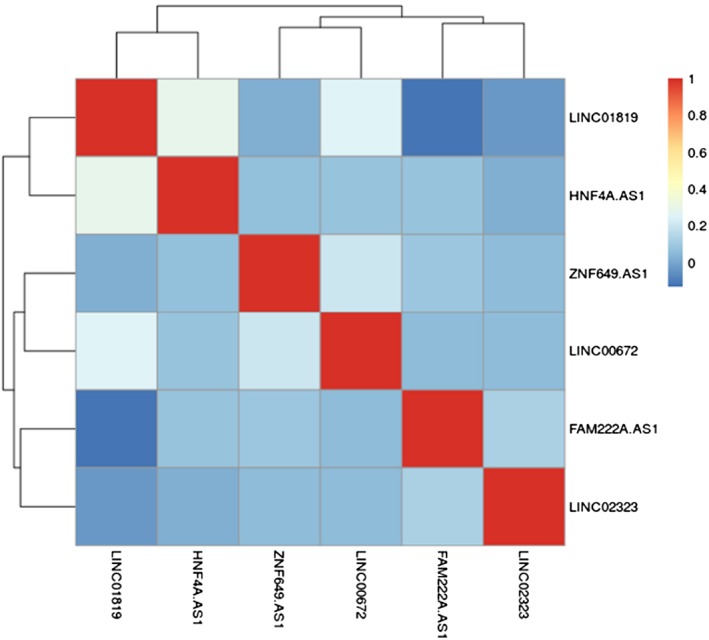
Correlation matrix of expression levels between six lncRNAs.
Evaluation of the prognostic performance for the six‐lncRNA signature
According to the risk score formula, the risk score was calculated for each patient in the training cohort. Next, patients were stratified into the low‐risk group and high‐risk group via the same median risk score as the cutoff point in the training, testing and entire sets. The relapse risk based on the six‐lncRNA signature was found to be significantly associated with N stage in the training cohort (Table 1). Kaplan‐Meier analysis indicated that there was a significant difference in RFS between the high‐ and low‐risk groups. The prognosis of the high‐risk group was significantly poorer compared with the low‐risk group (log‐rank test, all P < 0.001) (Fig 3). Meanwhile, the individual lncRNAs of the signature also showed large differences in RFS between the high‐ and low‐risk groups (Fig S1). Figure 4 illustrated the predictive potential of the six‐lncRNA signature using time‐dependent ROC curves. The AUCs of the signature for RFS prediction were larger than the TNM staging system (0.7059 vs. 0.6678 at three years, P = 0.0209; 0.7307 vs. 0.6111 at five years, P = 0.0468) in the entire TCGA cohort. When we combined the lncRNA signature with the TNM stage, the AUCs were further improved to 0.7474 at three years and 0.7670 at five years, respectively, with significant differences in comparison to TNM stage alone (P = 0.0006 and P = 0.0096). There was no significant difference in three‐ and five‐year RFS prediction between two methods by using the lncRNA signature and the combination model of the signature and TNM stage (P = 0.1331 and P = 0.2138, respectively). In the univariate and multivariate Cox regression analyses of RFS, the high‐risk group showed a 2.80‐fold increase in relapse risk (95% CI, 1.960–4.000, P = 1.46E‐08) compared with the low‐risk group, and the lncRNA signature was independent of traditional clinical risk factors (Table 3).
Figure 3.
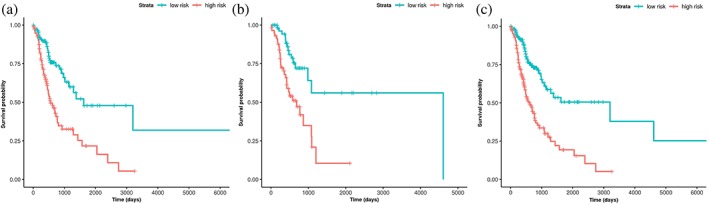
Kaplan‐Meier analysis of RFS in the training (a), testing (b) and entire (c) sets.
Figure 4.
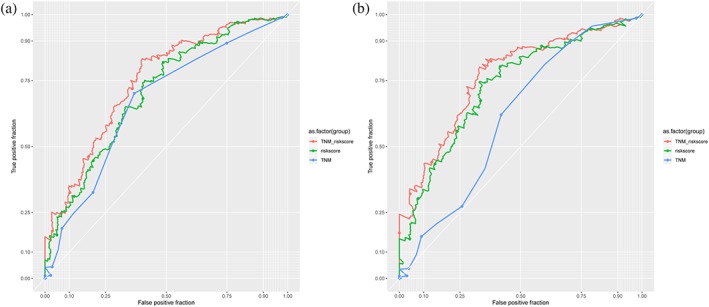
Time‐dependent ROC curves to compare the prognostic accuracy of the six‐IncRNA classifier with tumor stage in TCGA cohort. (a) AUCs at three years. (b) AUCs at five years.
Table 3.
Univariate and multivariate Cox proportional hazards regression analysis of factors associating with RFS in all 347 patients
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Label | HR | 95% CI | P‐value | HR | 95% CI | P‐value |
| Age | 1.010 | 0.991–1.020 | 0.389 | |||
| Gender | ||||||
| Male versus Female | 0.875 | 0.627–1.220 | 0.434 | |||
| Stage | ||||||
| II‐ IV versus I | 1.150 | 1.070–1.240 | 0.000 | |||
| T stage | ||||||
| T2‐4 versus T1 | 1.100 | 1.010–1.200 | 0.029 | 1.061 | 0.972–1.158 | 0.187 |
| N stage | ||||||
| N+ versus N0 | 1.240 | 1.060–1.450 | 0.008 | 1.174 | 0.986–1.397 | 0.071 |
| M stage | ||||||
| M1 versus M0 | 1.070 | 0.939–1.220 | 0.306 | |||
| lncRNA panel | ||||||
| High risk versus low risk | 2.800 | 1.960–4.000 | 0.000 | 2.724 | 1.907–3.893 | 0.000 |
Potential metastasis‐associated function of six lncRNAs
Figure 5a illustrates the expressions of six lncRNAs in four LUAD cell lines, indicating a relatively low expression level of all the six lncRNAs in A549 cells. Therefore, the lncRNA expression plasmids were transfected into A549 cells to over‐express these lncRNAs, respectively. The in vitro invasion assays showed that all these lncRNAs had a regulatory role in cell invasion ability, and the most obvious changes of the cell mobility were found in the LINC02323‐transfected cells (Fig 5b). This effect on cell migration and invasion was further determined in A549 and SPC‐A‐1 cells through both over‐expression and depletion of LINC02323 (Fig 5c).
Figure 5.
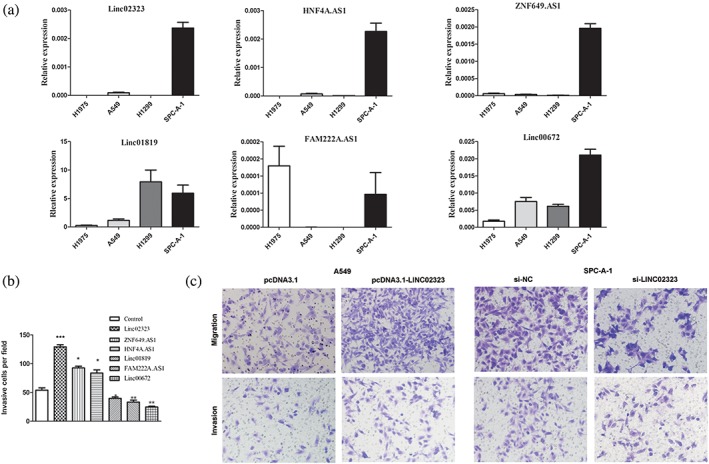
LincRNA expression patterns in LUAD cell lines. (a) Expression levels of six candidate lincRNAs in four LUAD cell lines as determined by qRT‐PCR. (b) Migration of A549 cells transfected with Linc02323, HNF4A.AS1, ZNF649.AS1, Linc01819, FAM222A.AS1, Linc00672 and control plasmids. The number of the invaded cells per field was counted after 24 hours of transfection. (c) Migration and invasion of A549 and SPC‐A‐1 cells with forced expression or siRNA knockdown of linc02323, respectively. The left two panels are the quantification of A549 cells that migrated through the pores of the membrane without the Matrigel (upper) and invaded through Matrigel‐coated membrane (lower). The right two panels represent SPC‐A‐1 cells that migrated or invaded across the Transwell membrane without (upper) or with Matrigel (lower). Error bars indicate the mean ± SD. *P < 0.05,**P < 0.01, ***P < 0.001.
Discussion
Increasing evidence shows that lncRNAs play a critical role in the metastasis and prognosis of various cancers.21 In the present study, we firstly screened differentially expressed metastasis‐associated lncRNAs using a TCGA LUAD cohort subset. Next, we constructed a metastasis‐associated lncRNA signature (LINC01819, ZNF649‐AS1, HNF4A‐AS1, FAM222A‐AS1, LINC02323 and LINC00672) for predicting RFS in LUAD patients, followed by validation of its clinical value. Additionally, we also determined the regulatory role of these lncRNAs in the metastasis of lung cancer.
A four‐gene prognostic signature, including one lncRNA gene, has been previously reported for LUAD patients, which performs well in stage I patients as well as EGFR‐mutant and wild‐type cohorts.9 Similarly, other studies have focused on characterizing gene signatures for prognosis of lung cancers.22, 23, 24 Until now, only a few lncRNA signatures have been characterized and proposed as biomarkers for lung cancer prognosis.25, 26, 27 In the current study, 735 lncRNAs were found differentially expressed between LUAD patients with and without metastasis, among which 43 lncRNAs were identified to be significantly associated with RFS of patients. Furthermore, six differentially expressed lncRNAs were obtained to construct a predictive model that demonstrated significant prognostic performance for relapse in LUAD patients. Unlike previous studies, our current study was the first report which investigated a metastasis‐associated lncRNA signature for the prognosis of LUAD.
We found that three lncRNAs, including LINC02323, ZNF649‐AS1 and HNF4A.AS1, were risk factors, while the other three lncRNAs, including LINC01819, FAM222A‐AS1 and LINC00672 were protective factors. High levels of risk factors and low levels of protective factors seemed to present independent negative prognostic factors. ZNF649‐AS1, the risk factor according to our analysis, is encoded by the antisense chain of ZNF649 that functions as a transcriptional repressor in mitogen‐activated protein kinase (MAPK) signaling pathway.28 MAPK pathway is known to be associated with tumor proliferation as well as metastasis.29, 30 Therefore, we speculated that ZNF649‐AS1 might be involved in the process of tumor metastasis and act as a oncogene in lung cancer. LINC00672, the protective factor in our analysis, has been consistently reported to contribute to p53 protein‐mediated gene suppression and promote endometrial cancer chemosensitivity.31 However, other lncRNAs, LINC01819, HNF4A‐AS1, FAM222A‐AS1 and LINC02323, were reported here for the first time in lung cancer.
The combined index of our six lncRNAs exhibited superior clinical significance in LUAD patients. Kaplan‐Meier analysis indicated that there was a significant difference in RFS between the high‐ and low‐risk groups. Time‐independent ROC analyses showed the superiority of this signature in predicting outcome of LUAD patients compared with the conventional TNM staging system. In order to improve the prognostic accuracy, we combined the six‐lncRNA signature with clinicopathological risk factors. However, there was no significant difference between the lncRNA signature and the combined model, indicating that our six‐lncRNA signature could yield reliable predictive ability. Univariate analysis indicated that the six‐lncRNA signature was closely associated with RFS, and the multivariate analysis revealed that it could predict the relapse of LUAD patients independently of traditional clinical risk factors.
In this study, we used TCGA data which provided a foundation for the systematic analysis of large‐scale lncRNA expression data and enabled us to perform this comprehensive study. Moreover, our in vitro transwell assays revealed that all the lncRNAs in our six‐lncRNA signature played a regulatory role during the migration and invasion of lung cancer cells. As expected, three lncRNAs, the risk factors in our analysis, promoted the invasive ability of lung cancer cells, while the other three as protective factors inhibited the cell mobility in vitro. These results further strengthened our hypothesis that the metastasis‐associated lncRNAs had the potential application in targeted therapy and prognosis of LUAD patients, and these lncRNAs also provided the primary rationale to develop the lncRNA signature for predicting prognosis in LUAD patients.
Collectively, we identified a unique metastasis‐associated lncRNA signature which was related to not only tumor metastasis but also prognosis prediction of relapse in LUAD patients. This signature was an effective prognostic marker for LUAD, and might aid in the development of targeted therapies as a target in metastatic cancer. In addition, further studies are required to comprehensively assess the exact contribution of these lncRNAs to tumor progression.
Disclosure
The authors declare that there are no conflicts of interest.
Supporting information
Table S1. Summary of 3483 differentially expressed lncRNAs.
Table S2. Sequences of specific primers.
Table S3. Identification of the Prognostic lncRNAs as candidate variables from the TCGA Cohort.
Figure S1. Kaplan–Meier survival plots for each individual IncRNA in TCGA cohort.
Acknowledgments
This work was supported by a grant from the Fundamental Research Funds of Shandong University (2018JC002); Taishan Scholar Program of Shandong Province; Shandong Technological Development Project (2016GSF201120, 2019GSF108054); Key Research and Development Project of Shandong Province (2019GSF108034); Natural Science Foundation of Shandong Province (ZR2017MH014).
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2. Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015; 16: e342–51. [DOI] [PubMed] [Google Scholar]
- 3. Campbell JD, Alexandrov A, Kim J et al Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016; 48: 607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fatica A, Bozzoni I. Long non‐coding RNAs: New players in cell differentiation and development. Nat Rev Genet 2014; 15: 7–21. [DOI] [PubMed] [Google Scholar]
- 5. Yin D, Lu X, Su J et al Long noncoding RNA AFAP1‐AS1 predicts a poor prognosis and regulates non‐small cell lung cancer cell proliferation by epigenetically repressing p21 expression. Mol Cancer 2018; 17: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang B, Zhang L, Cao Y et al Overexpression of lncRNA IGFBP4‐1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer 2017; 16: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jen J, Tang YA, Lu YH, Lin CC, Lai WW, Wang YC. Oct4 transcriptionally regulates the expression of long non‐coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol Cancer 2017; 16: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng F, Wang R, Zhang Y et al Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer 2017; 16: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shukla S, Evans JR, Malik R et al Development of a RNA‐Seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst 2017; 109: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie Y, Zhang Y, Du L et al Circulating long noncoding RNA act as potential novel biomarkers for diagnosis and prognosis of non‐small cell lung cancer. Mol Oncol 2018; 12: 648–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li RH, Chen M, Liu J et al Long noncoding RNA ATB promotes the epithelial‐mesenchymal transition by upregulating the miR‐200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis 2018; 9: 1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang PS, Chou CH, Lin CH et al A novel long non‐coding RNA linc‐ZNF469‐3 promotes lung metastasis through miR‐574‐5p‐ZEB1 axis in triple negative breast cancer. Oncogene 2018; 37: 4662–78. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Wang W, Xia P et al Identification of a five‐lncRNA signature for predicting the risk of tumor recurrence in patients with breast cancer. Int J Cancer 2018; 143: 2150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang L, Wang R, Fang L et al HCP5 is a SMAD3‐responsive long non‐coding RNA that promotes lung adenocarcinoma metastasis via miR‐203/SNAI axis. Theranostics 2019; 9: 2460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ge X, Li GY, Jiang L et al Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR‐203/30/SNAI axis. Oncogene 2019; 38: 3061–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang JX, Song W, Chen ZH et al Prognostic and predictive value of a microRNA signature in stage II colon cancer: A microRNA expression analysis. Lancet Oncol 2013; 14: 1295–306. [DOI] [PubMed] [Google Scholar]
- 17. Qu A, Wang W, Yang Y et al A serum piRNA signature as promising non‐invasive diagnostic and prognostic biomarkers for colorectal cancer. Cancer Manag Res 2019; 11: 3703–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qu A, Yang Y, Zhang X et al Development of a preoperative prediction nomogram for lymph node metastasis in colorectal cancer based on a novel serum miRNA signature and CT scans. EBioMedicine 2018; 37: 125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y, Qu A, Zhao R et al Genome‐wide identification of a novel miRNA‐based signature to predict recurrence in patients with gastric cancer. Mol Oncol 2018; 12: 2072–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heagerty PJ, Lumley T, Pepe MS. Time‐dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000; 56: 337–44. [DOI] [PubMed] [Google Scholar]
- 21. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016; 29: 452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu C, Li Y, Wei M, Zhao L, Yu Y, Li G. Identification of a novel glycolysis‐related gene signature that can predict the survival of patients with lung adenocarcinoma. Cell Cycle 2019; 18: 568–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qi L, Li T, Shi G et al An individualized gene expression signature for prediction of lung adenocarcinoma metastases. Mol Oncol 2017; 11: 1630–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen DT, Hsu YL, Fulp WJ et al Prognostic and predictive value of a malignancy‐risk gene signature in early‐stage non‐small cell lung cancer. J Natl Cancer Inst 2011; 103: 1859–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang L, Zhao H, Xu Y et al Systematic identification of lincRNA‐based prognostic biomarkers by integrating lincRNA expression and copy number variation in lung adenocarcinoma. Int J Cancer 2019; 144: 1723–34. [DOI] [PubMed] [Google Scholar]
- 26. Miao R, Ge C, Zhang X et al Combined eight‐long noncoding RNA signature: A new risk score predicting prognosis in elderly non‐small cell lung cancer patients. Aging (Albany NY) 2019; 11: 467–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Xie D, He Z, Zheng L. Integrated analysis reveals five potential ceRNA biomarkers in human lung adenocarcinoma. PeerJ 2019; 7: e6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang H, Yuan W, Wang Y et al ZNF649, a novel Kruppel type zinc‐finger protein, functions as a transcriptional suppressor. Biochem Biophys Res Commun 2005; 333: 206–15. [DOI] [PubMed] [Google Scholar]
- 29. Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: An update. Arch Toxicol 2015; 89: 867–82. [DOI] [PubMed] [Google Scholar]
- 30. Zhao T, Ren H, Li J et al LASP1 is a HIF1α target gene critical for metastasis of pancreatic cancer. Cancer Res 2015; 75: 111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li W, Li H, Zhang L et al Long non‐coding RNA LINC00672 contributes to p53 protein‐mediated gene suppression and promotes endometrial cancer chemosensitivity. J Biol Chem 2017; 292: 5801–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Summary of 3483 differentially expressed lncRNAs.
Table S2. Sequences of specific primers.
Table S3. Identification of the Prognostic lncRNAs as candidate variables from the TCGA Cohort.
Figure S1. Kaplan–Meier survival plots for each individual IncRNA in TCGA cohort.


