Abstract
Background
The development of biomarkers for the early detection of non‐small cell lung cancer (NSCLC) is clinically important. We have developed miRNA biomarkers in sputum and plasma, respectively, for NSCLC. Herein, we evaluate whether integrated analysis of the miRNAs across the different types of specimens could improve the early detection of NSCLC.
Methods
Using reverse transcription PCR, we determined expressions of two miRNAs (miRs‐31‐5p and 210‐3p) in sputum and three miRNAs (miRs‐21‐5p, 210‐3p, and 486‐5p) in plasma of a training cohort of 76 NSCLC patients and 72 cancer‐free smokers. The results were validated in a testing cohort of 56 NSCLC patients and 55 cancer‐free smokers.
Results
The panels of two sputum miRNAs and three plasma miRNAs had 65.8–75.0% sensitivities and 83.3–87.5% specificities for diagnosis of NSCLC in the training cohort. The individual sputum or plasma miRNA panel had a higher sensitivity for squamous cell carcinoma or adenocarcinoma of the lung, respectively. From the miRNAs, we optimized an integrated panel of biomarkers consisting of two sputum miRNAs (miRs‐31‐5p and 210‐3p) and one plasma miRNA (miR‐21‐5p) that had higher sensitivity (85.5%) and specificity (91.7%) for diagnosis of NSCLC compared with the individual panels alone. Furthermore, the performance of the integrated panel of biomarkers was independent of histology and stage of NSCLC, and patients' age, sex, and ethnicity. The performance of the integrated panel of biomarkers was confirmed in the testing cohort.
Conclusions
Integrating biomarkers across different body fluids would synergistically improve the early detection of NSCLC.
Key points
Lung cancer is a heterogeneous disease and develops from complex aberrations.
Integrating sputum and plasma miRNAs has higher accuracy than when they are used alone
Keywords: Biomarkers, diagnosis, lung cancer, microRNA, plasma, sputum
Introduction
A total of 85% of lung cancers are non‐small cell lung cancer (NSCLC), which mainly consists of two histological types: adenocarcinoma (AC) and squamous cell carcinoma (SCC).1 The National Lung Screening Trial (NLST) shows that low‐dose CT (LDCT) could detect NSCLC at the early stage and significantly reduce mortality.2 However, LDCT has a high false‐positive rate (or a low specificity) in the early detection of lung cancer. Therefore, the development of body fluid‐based biomarkers for precisely diagnosing lung cancer at an early stage is clinically important.
It is well accepted that molecular changes in epithelial cells collected from the normal‐appearing mainstem bronchus of smokers could be developed as biomarkers for NSCLC.3 Sputum contains bronchial epithelial cells from the lungs and lower respiratory tract. Therefore, examination of the exfoliated bronchial airway epithelial cells in sputum can identify the lung tumor‐related molecular alterations, and thus provide a noninvasive and specific tool for diagnosis of NSCLC.3 MicroRNAs (miRNAs) are a class of small noncoding RNAs of ∼22nt in length.4 Dysregulations of miRNAs can drive multiple processes of tumorigenesis by regulating cell cycle, apoptosis, and migration.4 For example, we have shown that downregulations of miR‐486‐5p contribute to the development of NSCLC by regulating Pim‐1 and ARHGAP5.5, 6, 7, 8 We and others have also demonstrated that aberrant miRNA expressions detected in sputum biologically reflect those in primary lung cancer.9, 10, 11, 12, 13, 14, 15 We have recently developed a panel of two sputum miRNA (miR‐31‐5p and miR‐210‐3p) biomarkers for NSCLC with 65% sensitivity and 89% specificity.16 Furthermore, efforts have been made to develop cell‐free (eg, plasma) biomarkers by detecting the circulating molecules directly released from lung tumors. Plasma miRNAs released from primary lung tumors or the circulating cancer cells might also provide potential biomarkers for NSCLC. We have developed a panel of three plasma miRNA (miRs‐21‐5p, 210‐3p, and 486‐5p) biomarkers that have 75% sensitivity and 85% specificity for diagnosis of NSCLC.17
Lung cancer is a heterogeneous disease and develops from complex molecular aberrations. The objective of this study was to evaluate whether integrated analysis of the miRNAs in sputum and plasma could improve the early detection of NSCLC.
Methods
Specimen collection and preparation
The Institutional Review Boards (IRB) at University of Maryland Baltimore approved this study. Written informed consent was obtained from all participants. Sputum and blood samples were collected from the participants before they had any treatment as previously described.7, 9, 16, 18, 19, 20, 21, 22, 23, 24, 25, 26 The preparation of airway bronchial epitheliums from sputum was performed using a protocol developed in previous studies.7, 9, 16, 18, 19, 20, 21, 22, 23, 25, 26 Plasma was prepared from blood by using the standard operating protocols developed by The NCI‐Early Detection Research Network.27
Quantitative reverse transcription PCR (qRT‐PCR) for determination of miRNA expression levels
RNA was extracted from the clinical specimens by using our established protocols.9, 16 The expressions of two miRNAs (miRs‐31‐5p and 210‐3p) in sputum, and three miRNAs (miRs‐21‐5p, 210‐3p, and 486‐5p) in plasma were determined by using Taqman miRNA assays (Applied Biosystems, Foster City, CA).7, 9, 16, 19, 20, 21, 28 The levels of miRNAs were measured by using a threshold cycle method with U6 as an internal control to normalize the expression of each gene.7, 9, 16, 21, 29, 30, 31 We included controls in each experiment: RNA extracted from a H358 NSCLC cell line (a positive control for RT, preamplification and ddPCR), genomic DNA (a positive control for genomic DNA detection), and nuclease‐free water (control for contamination). All experiments were repeated three times.
Statistical analysis
We expected that the study had ≥90% power at ≤1 false positives to detect miRNAs with fold change ≥2 between cancer patients and control subjects. To achieve the statistical criteria, ≥55 specimens from each group were required. Statistical analysis of RT‐qPCR data was done using Statistical Analysis System software version 6.12 (SAS Institute, Cary, NC). All P‐values were two‐sided, and a P‐value of <0.05 was considered statistically significant. Furthermore, contingency table and logistic regression analysis were applied to determine the associations between the expression levels of the miRNAs and both clinicopathologic and demographic characteristics of the cases and controls. In addition, we analyzed the significantly associated miRNAs by using logistic regression models with constrained parameters as least absolute shrinkage and selection operator (LASSO) to eliminate the less relevant variables.32 We estimated functions of the combined miRNA biomarkers by logistic regression with or without adjustment for known risk factors for NSCLC. Receiver‐operator characteristic (ROC) curve analysis was undertaken using expression level for each miRNA from cancer patients and cancer‐free controls by Analyze‐it software (Analyze‐it Software, Leeds, UK). We also generated a 95% confidence interval for the difference in the area under the ROCs (AUCs) by using the bootstrap. We established the optimal cutoff value by using the Youden index. The results of the training cohort were blindly validated in the testing cohort by using leave‐one‐out cross validation. Moreover, to compare different miRNA biomarker panels, we computed their AUCs to determine the sensitivity and specificity as previously described.17
Results
Patients
We enrolled patients between the ages of 55–80 who had ≥ a 30 pack‐year smoking history and were former smokers (quit within 15 years). Exclusion criteria included pregnancy, current pulmonary infection, surgery within six months, radiotherapy within one year, and life expectancy of <one year. A total of 132 NSCLC patients and 127 cancer‐free smokers were recruited. No difference of age, race, gender and smoking status was observed in the lung cancer cases versus cancer‐free smokers. The cases and controls were randomly split into two cohorts: a training cohort and a testing cohort. The training cohort comprised 76 lung cancer patients and 72 cancer‐free smokers. The training cohort was used to simultaneously analyze sputum and plasma miRNAs, and hence develop an integrated panel of miRNA biomarkers for NSCLC. The testing cohort included 56 lung cancer patients and 55 cancer‐free smokers and was used for blindly confirming the results generated from the training cohort. Detailed demographic and clinical characteristics of the two cohorts are shown in Tables 1 and 2.
Table 1.
Characteristics of NSCLC patients and cancer‐free smokers in a training cohort
| NSCLC cases (n = 76) | Controls (n = 72) | P‐value | |
|---|---|---|---|
| Age | 67.45 (SD 12.35) | 66.63 (SD 10.37) | 0.33 |
| Sex | 0.45 | ||
| Female | 27 | 25 | |
| Male | 49 | 37 | |
| Race | 0.43 | ||
| White | 55 | 52 | |
| African American | 21 | 20 | |
| Pack‐years (median) | 35.46 | 35.22 | 0.38 |
| Stage | |||
| Stage I | 25 | ||
| Stage II | 23 | ||
| Stage III–IV | 28 | ||
| Histological type | |||
| Adenocarcinoma | 43 | ||
| Squamous cell carcinoma | 33 |
NSCLC, non‐small cell lung cancer.
Table 2.
Characteristics of NSCLC patients and cancer‐free smokers in a testing cohort
| NSCLC cases (n = 56) | Controls (n = 55) | P‐value | |
|---|---|---|---|
| Age | 66.87 (SD 11.36) | 66.06 (SD 10.78) | 0.39 |
| Sex | 0.40 | ||
| Female | 20 | 19 | |
| Male | 36 | 36 | |
| Race | 0.45 | ||
| White | 41 | 39 | |
| African American | 15 | 16 | |
| Pack‐years (median) | 35.68 | 35.13 | 0.38 |
| Stage | |||
| Stage I | 18 | ||
| Stage II | 17 | ||
| Stage III–IV | 21 | ||
| Histological type | |||
| Adenocarcinoma | 31 | ||
| Squamous cell carcinoma | 25 |
NSCLC, non‐small cell lung cancer.
Individual sputum miRNAs and plasma miRNAs could distinguish lung cancer patients from cancer‐free controls
In the training cohort, the two sputum miRNAs (miRs‐31‐5p and 210‐3p) and three miRNAs (miRs‐21‐5p, 210‐3p, and 486‐5p) exhibited a higher expression level in NSCLC patients versus cancer‐free smokers (all P < 0.001) (Table SS1). Combined analysis of the two sputum miRNAs as a panel produced 0.82 AUC with 65.8% sensitivity and 87.5% specificity for diagnosis of NSCLC (Table 3). The expression level of both miR‐21‐5p and miR‐210‐3p in sputum was correlated to SCC (P < 0.05). Therefore, the sputum miRNAs showed a higher sensitivity for diagnosis of SCC compared with AC (78.8% vs. 55.8%, P = 0.02), while having the same specificity (87.5%). However, the sputum miRNAs did not have a specific relationship with stage of lung cancer, and patients' age, race, and gender (All P > 0.05), but smoking history (P = 0.04).
Table 3.
Comparison of the integrated panel of two sputum and one plasma miRNAs with individual panels of sputum and plasma miRNAs in a training cohort
| AUC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | ||
|---|---|---|---|---|
| Two sputum miRNAs | 0.816 (0.756–0.882) | 65.79% (54.01%– 76.29%) | 87.50% (77.59%– 94.12%) | |
| The three plasma miRNAs | 0.852 (0.779–0.902) | 75.00% (63.74%–84.23%) | 83.33% (72.70%–91.08%) | |
| Integrated 2 sputum and 1 plasma miRNAs | 0.913 (0.864–0.956) | 85.53% (75.58%– 92.55%) | 91.67% (82.74%– 96.88%) |
AUC, the area under receiver operating characteristic curve; CI, confidence interval.
The analysis of the three plasma miRNAs had an AUC of 0.85, producing 75.0% sensitivity and 83.3% specificity for diagnosis of NSCLC (Table 3). The expression levels of the plasma miRNAs were related with AC. As a result, combined use of the three plasma miRNAs created a higher sensitivity (81.4.8% vs. 66.7%, P = 0.03) and the same specificity (83.3%) for diagnosis of AC vs. SCC. The plasma biomarkers did not have relationship with stage of NSCLC, and patients' age, sex, ethnicity, and smoking history of the participants (All > 0.05).
Integrated analysis of two miRNA biomarkers and one plasma miRNA biomarker has a synergistic effect on the early detection of NSCLC
From the multiple sputum and plasma miRNA biomarkers, we used logistic regression models with constrained parameters as in LASSO to identify and optimize biomarkers. Two sputum miRNAs (miRs‐31‐5p and 210‐3p) and one plasma miRNA (miRNA‐21‐5p) were finally selected as the best and incorporated into an algorithm, producing 0.91 AUC for distinguishing lung cancer from cancer free subjects (Figure S1). U1 = 18.847 + 2.812 × log(miR − 205 − 5p/u6) − 5.608 × log(miR145/U6) + 5.386 × log(miR − 210/U6) − 7.734 × log(miR126/U6)
Integrated analysis of the three biomarkers by using this algorithm could produce a greater AUC than the individual sputum biomarker panel or plasma biomarker panel (0.91 vs 0.82 and 0.85, all P < 0.05) (Table 3). Using Youden's index, we set the optimal cutoff at 3.52. Integrated use of the two sputum miRNAs and one plasma miRNA produced higher sensitivity (85.5%) and specificity (91.7%) than did the sputum and plasma biomarkers (all P < 0.05) (Table 3). Furthermore, combined use of all the five biomarkers (two miRNA and three plasma biomarkers) did not produce higher sensitivity and specificity compared with the integrated panel of the three biomarkers (P > 0.05). In addition, the estimated Pearson correlation among levels of the three miRNAs was very low (all P > 0.05), suggesting that the integration had complementary classification for diagnosis of lung cancer. Moreover, in contrast to either the sputum or plasma miRNA biomarker panel used alone, integrated analysis of the three biomarkers across the different body fluids did not show a special association with histology of lung cancer. In addition, the integrated panel of biomarkers had no association with stage of lung cancer, and patients' age, race, and gender (all P > 0.05).
Validating the synergistic effect of the integrated panel of three miRNAs across the body fluids for diagnosis of NSCLC in the testing cohort
The two sputum miRNAs (miRs‐31‐5p and 210‐3p) and one plasma miRNA (miR‐21‐5p) were analyzed in the specimens of 56 lung cancer patients and 55 cancer‐free smokers. The integration of the three biomarkers across sputum and plasma showed higher sensitivity (83.9% vs. 64.3% and 75.0%) and specificity (90.9% vs. 87.3% and 83.6%) than did the individual sputum miRNA panel and plasma miRNA panel (all P < 0.05) (Fig 1). Furthermore, integrated analysis of the three biomarkers did not display a particular relationship with histological types and stage of NSCLC, and patients' age, race, and gender (all P > 0.05), supporting the synergistic value of this integrated panel of biomarkers for the early detection of lung cancer.
Figure 1.
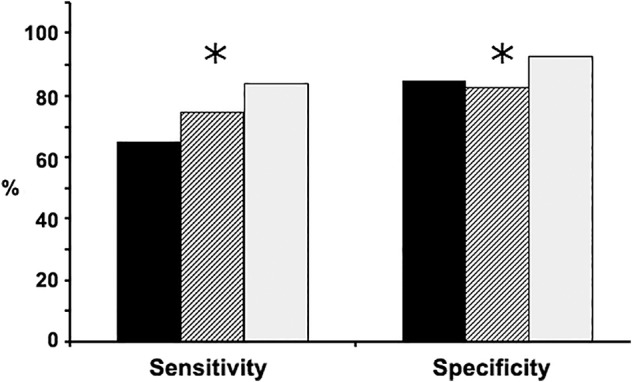
Compares the integrated panel of the three biomarkers with the individual sputum miRNA panel and plasma miRNA panel for lung cancer diagnosis in the testing cohort. The integration of the three biomarkers across sputum and plasma had higher sensitivity and specificity compared with the individual sputum miRNA panel and plasma miRNA panel. *P < 0.05.  Sputum biomarkers,
Sputum biomarkers,  plasma biomarkers,
plasma biomarkers,  integrated sputum and plasma biomarkers.
integrated sputum and plasma biomarkers.
Discussion
Most lung SCCs are located centrally, usually in the larger bronchi that join the trachea to the lung, whereas ACs often arise in peripheral lung tissue and originate from the smaller airways of the lungs. Sputum and plasma have different characteristics as surrogate material for molecular diagnosis of NSCLC. In particular, sputum miRNAs provide cell‐based biomarkers that are developed from molecular changes of the exfoliated bronchial epitheliums of large airways or main bronchial where SCC more commonly exists. In contrast, cell‐free plasma miRNA biomarkers are based on detecting the circulating molecules directly released from lung tumors and the floating cancer cells. Therefore, sputum biomarkers could be more sensitive for SCCs, whereas plasma biomarkers might be more sensitive for ACs. Given that altered miRNAs in sputum and plasmas contribute to lung tumorigenesis via the different mechanisms, integrating the miRNAs across the different body fluids may have a synergetic effect. Indeed, here we demonstrate that the combined analysis of the two sputum (cell‐based) miRNA biomarkers and one plasma (cell‐free circulating) miRNA biomarkers generates a better performance than did a single category of the miRNA biomarkers. Furthermore, no statistical association exists among the two sputum miRNAs and one plasma miRNA, further supporting that the integration of the two types of miRNA biomarkers might have a synergistic value. Through a complementary manner, integrating sputum miRNAs with plasma miRNAs could diagnose lung cancer, irrespective of the histology and stage. This finding is consistent with our previous discovery,33 in which combined analysis of miRNA expressions in circulating peripheral blood mononucleated cells and sputum might have a synergistic effect for lung cancer detection. Moreover, integrated analysis of the two sputum and one plasma biomarkers does not show a statistical relationship with patients' age, race, and gender. The findings would be clinically important if the integrated biomarker panel is used for identifying early stage lung cancer.
Dysregulations of miRs‐21‐5p, 31‐5p, and 210‐3p have been reported to be associated with a variety of malignancies, including NSCLC.34, 35, 36 For example, the miR‐21 family has an important oncogenic role, whose overexpression contributes to tumor growth and metastasis, and is associated with poor prognosis and chemosensitive tumors.34 Furthermore, anti‐miR‐21 has been demonstrated to suppress cell growth of breast cancer through downregulation of the antiapoptotic factor, B‐cell lymphoma 2 (Bcl‐2). miR‐210‐3p has been reported to stimulate a hypoxic phenotype and upsurge radioresistance in NSCLCs.35 Hypoxia‐induced miR‐210‐3p can regulate tumor cell susceptibility to cytolytic T‐lymphocyte‐mediated lysis by a mechanism involving its downstream targets PTPN1, HOXA1, and TP53I11. Aberrant miR‐210‐3p expression in body fluids has been shown to assist in the diagnosis of several types of malignancies, including lung cancer.37 miR‐31‐5p downregulation in breast cancer is related to tumor metastases, whereas elevated miR‐31‐5p expression in colorectal cancer may be associated with late stage tumors.36 miR‐31‐5p dysregulation endorses KRAS mutation‐driven NSCLC, and hence plays a key role in lung tumorigenesis.38 Our current observations that the elevated expression levels of the three miRNAs in sputum and plasma of NSCLC patients further support their important roles in lung carcinogenesis.
The study may have some limitations. First, clinically useful biomarkers should have very high sensitivity and specificity for diagnosis of lung cancer. Integrated use of three miRNAs has a sensitivity of 85% and a specificity of 91%, which are still not sufficient to be used in the clinics for the early detection of lung cancer. We are aiming to identify new lung tumor‐associated miRNAs of the exfoliated bronchial epitheliums of airway in sputum by using whole genome next‐generation sequencing, which may provide additional biomarkers to be added in the integrated approach to more precisely diagnose lung cancer. Second, using LDCT for the early detection of lung cancer can reduce the mortality.39 However, LDCT always produces over‐diagnosis or a false positive rate.39 It would be interesting to evaluate whether the integrated panel of miRNA biomarkers could complement LDCT for the early detection of lung cancer by specifically reducing its over‐diagnosis. Third, small cell lung cancer (SCLC) accounts for approximately 15% of all lung cancers.1 However, specimens from SCLC patients were not available in this current study. Diagnostic potential of the biomarkers for the early detection of SCLC remains unknown. In our ongoing study, we are collecting sputum and plasma samples from patients diagnosed with SCLC to evaluate whether these biomarkers could be useful for the diagnosis of SCLC.
In conclusion, integrating sputum and plasma miRNA biomarkers has a synergistic value, and hence presents a potential approach for the early detection of NSCLC. Nevertheless, a large multicenter trial is required to confirm the integrated method before it can be translated in laboratory settings.
Disclosure
The authors declare no conflict of interest.
Supporting information
Table S1 Expression levels of the sputum miRNAs and plasma miRNAs in NSCLC patients versus cancer‐free controls of the training cohort.
Figure S1 The receiver‐operator characteristic (ROC) curve of sputum and plasma miRNA biomarkers for lung cancer diagnosis. Two sputum miRNAs (miRs‐21‐5p and 210‐3p) and one plasma miRNA (miRNA‐31‐5p) were selected as the best ones and incorporated into an algorithm: P = ex/ (1 + ex), where x = 6.62 + 1.84 × log (miR‐21‐5p)‐2.6 × log (miR‐21‐3p) ‐2.36 × log (miR‐31‐5p). The algorithm produced the area under the ROC (AUC) of 0.91 for distinguishing lung cancer cases from controls.
Acknowledgments
This work was supported in part by the Geaton and JoAnn DeCesaris Family Foundation and University of Maryland Marlene & Stewart Greenebaum Comprehensive Cancer Center Pilot Grant Program (F.J.).
References
- 1. Torre LA, Bray F, Siegel RL et al Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87‐108. [DOI] [PubMed] [Google Scholar]
- 2. Aberle DR, Adams AM, Berg CD et al Reduced lung‐cancer mortality with low‐dose computed tomographic screening. N Engl J Med 2011; 365: 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belinsky SA, Liechty KC, Gentry FD et al Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high‐risk cohort. Cancer Res 2006; 66: 3338–44. [DOI] [PubMed] [Google Scholar]
- 4. Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell 2005; 122: 6–7. [DOI] [PubMed] [Google Scholar]
- 5. Wang J, Tian X, Han R et al Downregulation of miR‐486‐5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene 2014; 33: 1181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pang W, Tian X, Bai F et al Pim‐1 kinase is a target of miR‐486‐5p and eukaryotic translation initiation factor 4E, and plays a critical role in lung cancer. Mol Cancer 2014; 13: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol 2010; 23: 1157–64. [DOI] [PubMed] [Google Scholar]
- 8. Belinsky SA, Leng S, Wu G et al Gene methylation biomarkers in sputum and plasma as predictors for lung cancer recurrence. Cancer Prev Res (Phila) 2017; 10: 635–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xie Y, Todd NW, Liu Z et al Altered miRNA expression in sputum for diagnosis of non‐small cell lung cancer. Lung Cancer 2010; 67: 170–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fusco N, Fumagalli C, Guerini‐Rocco E. Looking for sputum biomarkers in lung cancer secondary prevention: Where are we now? J Thorac Dis 2017; 9: 4277–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bagheri A, Khorshid HRK, Tavallaie M et al A panel of noncoding RNAs in non‐small‐cell lung cancer. J Cell Biochem 2018; 23:35‐8. [DOI] [PubMed] [Google Scholar]
- 12. He RQ, Cen WL, Cen JM et al Clinical significance of miR‐210 and its prospective Signaling pathways in non‐small cell lung cancer: Evidence from gene expression omnibus and the cancer genome atlas data mining with 2763 samples and validation via real‐time quantitative PCR. Cell Physiol Biochem 2018; 46: 925–52. [DOI] [PubMed] [Google Scholar]
- 13. Bagheri A, Khorram Khorshid HR, Mowla SJ et al Altered miR‐223 expression in sputum for diagnosis of non‐small cell lung cancer. Avicenna J Med Biotechnol 2017; 9: 189–95. [PMC free article] [PubMed] [Google Scholar]
- 14. Razzak R, Bedard EL, Kim JO et al MicroRNA expression profiling of sputum for the detection of early and locally advanced non‐small‐cell lung cancer: A prospective case‐control study. Curr Oncol 2016; 23: e86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim JO, Gazala S, Razzak R et al Non‐small cell lung cancer detection using microRNA expression profiling of bronchoalveolar lavage fluid and sputum. Anticancer Res 2015; 35: 1873–80. [PubMed] [Google Scholar]
- 16. Shen J, Liao J, Guarnera MA et al Analysis of MicroRNAs in sputum to improve computed tomography for lung cancer diagnosis. J Thorac Oncol 2014; 9: 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shen J, Liu Z, Todd NW et al Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer 2011; 11: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohen JD, Li L, Wang Y et al Detection and localization of surgically resectable cancers with a multi‐analyte blood test. Science 2018; 359: 926–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu L, Shen J, Mannoor K, Guarnera M, Jiang F. Identification of ENO1 as a potential sputum biomarker for early‐stage lung cancer by shotgun proteomics. Clin Lung Cancer 2014; 15: 372–378.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li N, Ma J, Guarnera MA, Fang H, Cai L, Jiang F. Digital PCR quantification of miRNAs in sputum for diagnosis of lung cancer. J Cancer Res Clin Oncol 2014; 140: 145–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anjuman N, Li N, Guarnera M, Stass SA, Jiang F. Evaluation of lung flute in sputum samples for molecular analysis of lung cancer. Clin Transl Med 2013; 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu L, Todd NW, Xing L et al Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer 2010; 127: 2870–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang F, Todd NW, Li R, Zhang H, Fang H, Stass SA. A panel of sputum‐based genomic marker for early detection of lung cancer. Cancer Prev Res (Phila) 2010; 3: 1571–8. [DOI] [PubMed] [Google Scholar]
- 24. Jiang F, Todd NW, Qiu Q, Liu Z, Katz RL, Stass SA. Combined genetic analysis of sputum and computed tomography for noninvasive diagnosis of non‐small‐cell lung cancer. Lung Cancer 2009; 66: 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qiu Q, Todd NW, Li R et al Magnetic enrichment of bronchial epithelial cells from sputum for lung cancer diagnosis. Cancer 2008; 114: 275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li R, Todd NW, Qiu Q et al Genetic deletions in sputum as diagnostic markers for early detection of stage I non‐small cell lung cancer. Clin Cancer Res 2007; 13: 482–7. [DOI] [PubMed] [Google Scholar]
- 27. Tuck MK, Chan DW, Chia D et al Standard operating procedures for serum and plasma collection: Early detection research network consensus statement standard operating procedure integration working group. J Proteome Res 2009; 8: 113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma J, Lin Y, Zhan M, Mann DL, Stass SA, Jiang F. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab Invest 2015; 95: 1197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xing L, Su J, Guarnera MA et al Sputum microRNA biomarkers for identifying lung cancer in indeterminate solitary pulmonary nodules. Clin Cancer Res 2015; 21: 484–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su J, Liao J, Gao L et al Analysis of small nucleolar RNAs in sputum for lung cancer diagnosis. Oncotarget 2015; 7: 5131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen J, Todd NW, Zhang H et al Plasma microRNAs as potential biomarkers for non‐small‐cell lung cancer. Lab Invest 2011; 91: 579–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su Y, Guarnera MA, Fang H, Jiang F. Small non‐coding RNA biomarkers in sputum for lung cancer diagnosis. Mol Cancer 2016; 36: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Su J, Leng Q, Lin Y et al Integrating circulating immunological and sputum biomarkers for the early detection of lung cancer. Biomark Cancer 2018; 10: 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA‐21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008; 18: 350–9. [DOI] [PubMed] [Google Scholar]
- 35. Grosso S, Doyen J, Parks SK et al MiR‐210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis 2013; 4: e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang S, Hu J, Zhang D, Li J, Fei Q, Sun Y. Prognostic role of microRNA‐31 in various cancers: A meta‐analysis. Tumour Biol 2014; 35: 11639–45. [DOI] [PubMed] [Google Scholar]
- 37. Chang W, Lee CY, Park JH et al Survival of hypoxic human mesenchymal stem cells is enhanced by a positive feedback loop involving miR‐210 and hypoxia‐inducible factor 1. J Vet Sci 2013; 14: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edmonds MD, Boyd KL, Moyo T et al MicroRNA‐31 initiates lung tumorigenesis and promotes mutant KRAS‐driven lung cancer. J Clin Invest 2016; 126: 349–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patz EF Jr, Pinsky P, Gatsonis C et al Overdiagnosis in low‐dose computed tomography screening for lung cancer. JAMA Intern Med 2014; 174: 269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Expression levels of the sputum miRNAs and plasma miRNAs in NSCLC patients versus cancer‐free controls of the training cohort.
Figure S1 The receiver‐operator characteristic (ROC) curve of sputum and plasma miRNA biomarkers for lung cancer diagnosis. Two sputum miRNAs (miRs‐21‐5p and 210‐3p) and one plasma miRNA (miRNA‐31‐5p) were selected as the best ones and incorporated into an algorithm: P = ex/ (1 + ex), where x = 6.62 + 1.84 × log (miR‐21‐5p)‐2.6 × log (miR‐21‐3p) ‐2.36 × log (miR‐31‐5p). The algorithm produced the area under the ROC (AUC) of 0.91 for distinguishing lung cancer cases from controls.


