Abstract
Liquid biopsies are not used in practice for hepatocellular carcinoma (HCC). Epi proColon is the first commercial blood‐based test for colorectal cancer screening based on methylated DNA testing of the septin 9 gene (SEPT9). However, Epi proColon has some disadvantages, including the requirement of a large amount of blood and lack of quantitative performance. Therefore, we previously developed a novel liquid biopsy test that can quantitatively detect even a single copy of methylated SEPT9 in a small amount of DNA. In the current study, we evaluated the application potential of this assay for diagnosing HCC. Study subjects included 80 healthy volunteers, 45 patients with chronic liver disease (CLD) without HCC, and 136 patients with HCC (stage 0, 12; stage A, 50; stage B, 31; stage C, 41; and stage D, 2), according to the Barcelona Clinic Liver Cancer staging system. For the assay, DNA was treated with methylation‐sensitive restriction enzymes in two steps, followed by multiplex droplet digital polymerase chain reaction. The median copy number of methylated SEPT9 was 0.0, 2.0, and 6.4 in the healthy control, CLD, and HCC groups, respectively, with significant differences among the groups (HCC vs. healthy control, P < 0.001; HCC vs. CLD, P = 0.002; CLD vs. healthy control, P = 0.008). Assay sensitivity and specificity were 63.2% and 90.0%, respectively (cutoff value, 4.6 copies), in detecting HCC when compared with healthy subjects. The positive rate of methylated SEPT9 increased with HCC progression (stage 0, 41.7%; stage A, 58.0%; stage B, 61.3%; stage C, 75.6%; and stage D, 100%). Conclusion: We developed a sensitive methylated SEPT9 assay that might serve as a liquid biopsy test for diagnosing HCC.
We showed that a novel liquid biopsy test targeting methylated SEPT9 is useful for diagnosing HCC.

Abbreviations
- AFP
alpha‐fetoprotein
- AUC
area under the curve
- BCLC
Barcelona Clinic Liver Cancer
- CLD
chronic liver disease
- CORD
combined restriction digital polymerase chain reaction
- ctDNA
circulating tumor DNA
- ddPCR
droplet digital PCR
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- hTERT
human telomerase reverse transcriptase
- NBNC
non‐B non‐C
- PCR
polymerase chain reaction
- SEPT9
septin 9 gene
Hepatocellular carcinoma (HCC) is a common malignant disease and the fourth leading cause of cancer‐related deaths worldwide.1 The absolute number of primary cancer cases, of which HCC accounts for 75% to 85%, is expected to increase in the majority of 30 countries worldwide by 2030.2 To detect HCC at an early stage, international guidelines from the United States, Europe, Asia, and Japan advise HCC surveillance of at‐risk populations.3, 4, 5, 6 However, tumor biomarkers for the early detection of HCC are still lacking.4, 6 Because early screening of HCC patients is reportedly related to favorable survival,7, 8 a novel screening method for patients with increased risk for HCC is required.
Liquid biopsy has been suggested as a novel diagnostic tool, and circulating tumor cells and circulating tumor DNA (ctDNA) have been recognized as potential targets in liquid biopsies. Methylated ctDNA is among the most intensively investigated liquid biopsy targets. Because methylation changes in ctDNA occur early in carcinogenesis, numerous studies have evaluated the diagnostic performance of methylated genes, including cyclin‐dependent kinase inhibitor 2A (CDKN2A), Ras association domain family 1 isoform A (RASSF1A), and glutathione S‐transferase pi 1 (GSTP1), in patients with HCC.9, 10, 11 However, the clinical usefulness of these methylated ctDNA tests in HCC remains limited.
Epi proColon (Epigenomics AG, Berlin, Germany), which was approved by the US Food and Drug Administration (FDA) in 2016, was the first commercial blood‐based test for colorectal cancer screening based on methylated DNA testing of the septin 9 gene (SEPT9).12 There is only one report on the efficacy of Epi proColon in HCC patients.13 However, Epi proColon has some disadvantages. First, a large amount of blood plasma (> 3.5 mL) is required, because the amount of cancer‐specific DNA in blood is very small, and conventional DNA methylation assays require bisulfite treatment of DNA, which causes DNA degradation and loss.14, 15 Second, this test does not have quantitative performance, because it is based on real‐time polymerase chain reaction (PCR) plus/minus assay. To overcome these problems, we previously developed a new assay, the combined restriction digital PCR (CORD) assay, which circumvents the need for bisulfite treatment and methylated DNA immunoprecipitation. This assay is 100‐plus times more sensitive than the conventional bisulfide‐based methylation assay and thus overcomes the issue of limited input DNA. The CORD assay enables quantitative detection of even one copy of a methylated gene in a small DNA sample, without the need for DNA bisulfite treatment.16 The most unique feature of this novel assay is that it uses three methylation‐sensitive restriction enzymes in combination with droplet digital PCR (ddPCR). We have reported the performance of the serum CORD assay for methylated SEPT9 in patients with colorectal neoplasia, including cancer.17 In this study, we evaluated the potential application of this assay for methylated SEPT9 detection in HCC patients.
Experimental Procedures
This study was conducted in compliance with the ethical principles of the Declaration of Helsinki. The study protocol was approved by the institutional review boards of Yamaguchi University Hospital, Sentohiru Hospital, and Ajisu Kyoritsu Hospital (H28‐124 and H17‐83).
Serum and Tissue Samples
We enrolled 261 participants (Fig. 1) and prospectively and retrospectively collected 221 and 40 sera, respectively, between March 2015 and August 2018 at Yamaguchi University Hospital, Sentohiru Hospital, and Ajisu Kyoritsu Hospital. The subjects consisted of 80 healthy volunteers who had neither colorectal neoplasia as determined by colonoscopy nor chronic liver disease (CLD) (healthy control group), 45 patients with CLD without HCC (CLD group), and 136 patients with HCC (HCC group). Prospective data were obtained from each patient and healthy volunteer. Blood samples in the healthy control group were collected prior to bowel preparation for colonoscopy. Patients in the CLD group who showed no evidence of HCC by imaging modalities within 3 months of blood sampling and 6 months later were recruited, and blood samples in the HCC group were obtained immediately before HCC treatments. HCC was diagnosed based on Japanese guidelines.5 We assessed HCC tumor stage using the Barcelona Clinic Liver Cancer (BCLC) staging system.18 A total of 35 frozen tumor tissues and 34 matched frozen nontumor tissues (one sample was lost) were collected from 35 patients who underwent surgical resection of HCC.
Figure 1.
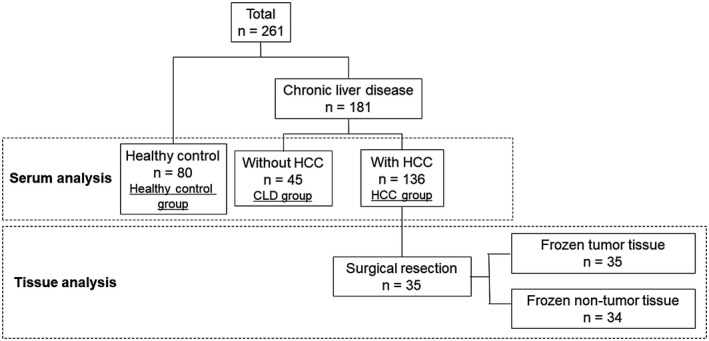
Study design. Of the 261 enrolled participants, 80 were healthy volunteers, 45 were patients with CLD without HCC, and 136 were patients with HCC. A total of 35 patients in the HCC group underwent surgical resection of HCC.
Sample Preparation and DNA Extraction
Samples were stored at −80°C until DNA extraction. Serum samples were thawed, and 0.4 mL of each sample was used for DNA extraction with the MagNA Pure Compact Nucleic Acid Isolation Kit I (Roche, Tokyo, Japan) according to the manufacturer's instructions. DNA was eluted in 50 µL of elution buffer. Tissue samples were thawed, and DNA was isolated using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). DNA was quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Tokyo, Japan).
CORD Assay of Methylated SEPT9
DNA samples were subjected to a CORD assay consisting of two‐step treatments with three methylation‐sensitive restriction enzymes, followed by multiplex ddPCR.16 In the first enzyme treatment step, 10 µL of circulating cell‐free DNA (an amount of DNA equivalent to the amount in 80 μL of serum) or 10 µL of 10 ng/µL tissue DNA was digested at 37°C for 16 hours by addition of 1 µL of GeneAmp 10× PCR Buffer II, 1 µL of 25 mmol/L magnesium chloride (MgCl2), 10 units of HhaI, 10 units of HpaII, and 20 units of exonuclease 1 (Exo1) (all from Thermo Fisher Scientific, Tokyo, Japan). Exo1 was added to eliminate single‐stranded DNA that would escape digestion by the restriction enzymes and to avoid PCR amplification of the undigested fraction.19 In the second step, DNA was further digested for 16 hours at 60°C using 10 units of BstUI (New England Biolabs, Hitchin, United Kingdom). After the restriction reaction was completed, the mixture was heated at 98°C for 10 minutes. SEPT9 has three recognition sites for the methylation‐sensitive enzymes HhaI and BstUI. When all three sites are methylated, the target DNA escapes digestion by these enzymes and is amplified by PCR. The human telomerase reverse transcriptase gene (hTERT) has no recognition sites for theses enzymes; therefore, hTERT is amplified by PCR when human DNA is present in the template DNA.
Absolute copy numbers of hTERT and methylated SEPT9 were determined by multiplex ddPCR. The 20‐µL PCR mixture consisted of 8 µL of enzyme‐treated serum cell‐free DNA (an amount of DNA equivalent to the amount in 0.04 mL serum) or 2 µL of enzyme‐treated tissue DNA, 1× ddPCR Supermix for Probes (Bio‐Rad, Tokyo, Japan), 0.25 µmol/L of each primer for a given target gene and an internal control, and 0.125 µmol/L of each probe for the target gene and internal control. We designed SEPT9 primers and probes within the CpG island 3 region of SEPT9 containing transcription start site of SEPT9 transcript variant 2, which is differentially methylated in colorectal cancer and is targeted by Epi proColon.20, 21 The primer and probe sequences for SEPT9 were as follows: forward primer, 5′GCCCACCAGCCATCATGT‐3′; reverse primer, 5′‐GTCCGAAATGATCCCATCCA‐3′; and probe, 5′‐FAM‐CCGCGGTCAACGC‐MGB‐3′. The PCR amplicon length was 62 base pairs (bp) (75,369,566‐75,369,627 on chromosome 17; human assembly Genome Reference Consortium human genome [build 37]/human genome 19 [GRCh37/hg19]). The primer and probe sequences for hTERT were as follows: forward primer, 5′GGGTCCTCGCCTGTGTACAG‐3′; reverse primer, 5′‐CCTGGGAGCTCTGGGAATTT‐3′; and probe, 5′‐VIC‐CACACCTTTGGTCACTC‐MGB‐3′.16 The amplicon length was 60 bp (1,253,375‐1,253,434 on chromosome 5). Droplet generation was carried out by an automated droplet generator (Bio‐Rad) and followed by PCR. PCR cycles included preheating at 95°C for 10 minutes, followed by 40 cycles of denaturation at 94°C for 30 seconds and annealing at 56°C for 60 seconds and final heating at 98°C for 10 minutes. After amplification, the PCR plate was transferred to a QX100 droplet reader (Bio‐Rad), and fluorescence amplitude data were obtained using QuantaSoft software (Bio‐Rad).
Alpha‐Fetoprotein Assay
Alpha‐fetoprotein (AFP) serum levels were measured using a LiBASys automated immunologic analyzer (Wako Pure Chemical Industries, Ltd., Osaka, Japan).
Statistical Analysis
Continuous variables are presented as the mean ± SD or median (interquartile range) and were compared using a paired or unpaired t test. Categorical variables were evaluated using the chi‐squared test or Fisher's exact test. Statistical significance was defined as a P value less than 0.05. The optimal cutoff value in receiver operating characteristic (ROC) analysis was determined by the Youden index. All analyses were performed using the JMP software package v13.0 (SAS Institute, Cary, NC).
Results
Patient Characteristics
Table 1 summarizes the clinical patient characteristics. The HCC group included 98 males (72%), with a mean age of 72.4 years. Etiology of HCC was hepatitis B virus (HBV) in 18 (13%), hepatitis C virus (HCV) in 58 (43%), and non‐B non‐C (NBNC) in 60 patients (44%). HCC tumor stage was 0, A, B, C, and D in 12 (8.8%), 50 (36.8%), 31 (22.8%), 41 (30.1%), and 2 patients (1.5%), respectively. Median tumor size and number were 3.2 cm and 1, respectively. A total of 33 patients (24%) had macrovascular invasion, and 14 (10%) showed extrahepatic spread. The CLD group included 22 males (49%), with a mean age of 69 years. Etiology was HBV in 12 (27%), HCV in 18 (40%), and NBNC in 15 (33%) patients.
Table 1.
Clinical Patient Characteristics in Serum Samples
| Healthy Control Group (n = 80) | CLD Group (n = 45) | HCC Group (n = 136) | |
|---|---|---|---|
| Age | 50.1 ± 8.5 | 69.0 ± 10.4 | 72.4 ± 8.5 |
| Gender, male/female | 38/42 | 22/23 | 98/38 |
| Etiology, HBV/HCV/NBNC | N/A | 12/18/15 | 18/58/60 |
| CH/LC (CTP grade A/B/C) | N/A | 12/33 (20/9/4) | 68/68 (51/17/0) |
| BCLC stage 0/A/B/C/D | N/A | N/A | 12/50/31/41/2 |
| Tumor size, cm | N/A | N/A | 3.2 (2.0‐5.75) |
| Tumor number | N/A | N/A | 1 (1‐3) |
| Macrovascular invasion +/− | N/A | N/A | 33/103 |
| Extrahepatic spread +/− | N/A | N/A | 14/122 |
Abbreviations: CH, chronic hepatitis; CTP, Child‐Turcotte‐Pugh; LC, liver cirrhosis; N/A; not applicable.
Basic Performance Evaluation of the CORD Assay
For basic performance evaluation of the CORD assay detecting hypermethylated cancer‐derived DNA against a background of blood‐derived DNA,17 we spiked EpiScope methylated HCT116 genomic DNA (control DNA for methylation of SEPT9; Takara Bio, Inc., Japan) at ratios of 100%, 50%, 10%, 5%, 1.1%, 0.11%, and 0% into DNA extracted from leukocyte DNA (control DNA for nonmethylated SEPT9) and measured the methylation level of SEPT9 in each sample. The CORD assay can quantify copy numbers of methylated SEPT9 from 6.25 pg of control methylated DNA (approximately two haploid genome copies) in a background of 5,625 pg of control nonmethylated DNA (Supporting Fig. S1).
Methylated SEPT9 Assay of Serum Samples
The median copy number of methylated SEPT9 was 0.0 (0.0‐3.4), 2.0 (0.0‐8.0), and 6.4 (2.6‐13.3) in the healthy control, CLD, and HCC groups, respectively, with significant differences among the groups (HCC group vs. healthy control group, P < 0.001; HCC group vs. CLD group, P = 0.002; CLD group vs. healthy control group, P = 0.008) (Fig. 2A). We performed ROC curve analysis to set an optimal cutoff value for distinguishing the healthy control group from the HCC group. When we set 4.6 copies of methylated SEPT9 as a provisional cutoff value, the area under the curve (AUC) was 0.81, with a sensitivity and specificity of 63.2% and 90.0%, respectively (Fig. 2B). On the other hand, assay sensitivity and specificity were 62.5% and 71.7%, respectively (cutoff value, 4.8 copies), in detecting HCC when compared with patients with CLD without HCC (data not shown). Therefore, 4.6 copies of methylated SEPT9 was set as the optimal cutoff value to identify methylation‐positive patients with HCC in the following analyses.
Figure 2.
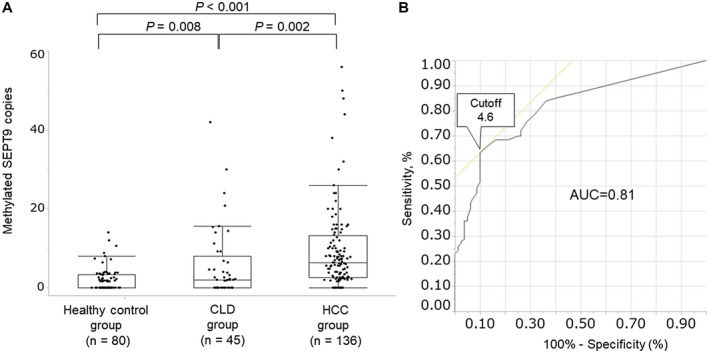
Serum methylated SEPT9 assay. (A) Methylated SEPT9 copy numbers in each of the study groups. The median copy number of methylated SEPT9 was 0.0 (0.0‐3.4), 2.0 (0.0‐8.0), and 6.4 (2.6‐13.3) in the healthy control group, CLD group, and HCC group, respectively. (B) AUC of methylated SEPT9. When we set 4.6 copies of methylated SEPT9 as a provisional cutoff value, the AUC was 0.81, with a sensitivity and specificity of 63.2% and 90.0%, respectively.
Correlation Between HCC Tumor Stage and Serum Methylated SEPT9 Level
The correlation between HCC tumor stage according to the BCLC staging system18 and methylated SEPT9 copy number is shown in Fig. 3. The copy number of methylated SEPT9 increased with HCC progression. The median copy number was 2.1, 5.6, 6.6, 9.2, and 53.7 in stage 0, A, B, C, and D, respectively (HCC group stage A, B, and C vs. healthy control group, P < 0.001, P < 0.001 and P < 0.001, respectively; HCC group stage C vs. CLD group, HCC group stage 0, A and B, P = 0.002, P = 0.002, P = 0.003 and P = 0.008, respectively) (Fig. 3A). The positive rate of methylated SEPT9 (cutoff ≥ 4.6 copies) increased from 41.7% to 100% with HCC progression (stage 0, A, B, C, and D, 41.7%, 58.0%, 61.3%, 75.6% and 100%, respectively) (Fig. 3B). There were significant differences in background liver disease, tumor size and number, and macrovascular invasion between methylated SEPT9 < 4.6 copies and ≥ 4.6 copies. In addition, the positive rate of methylated SEPT9 was 66.7% (40 of 60), 55.6% (10 of 18), and 62.1% (36 of 58) in patients with NBNC‐HCC, HBV‐related HCC, and HCV‐related HCC, respectively, with no significant difference among them (P = 0.672) (Supporting Table S1).
Figure 3.
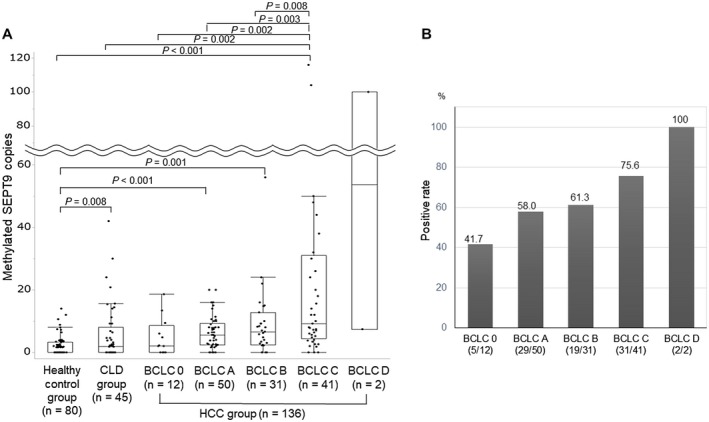
Correlation between HCC stage according to the BCLC staging system and serum methylated SEPT9. (A) The median copy number was 2.1, 5.6, 6.6, 9.2, and 53.7 in stage 0, A, B, C, and D, respectively. (B) The positive rate of serum methylated SEPT9 (cutoff value = 4.6 copies) increased from 41.7% to 100% with HCC progression.
Combination of Tumor Marker and Serum CORD Assay of Methylated SEPT9
We set 20 ng/mL and 4.6 copies as cutoff values for AFP and methylated SEPT9, respectively. The positive rates for both methods, AFP alone, and methylated SEPT9 alone were 27.2% (37 of 136), 11.0% (15 of 136), and 36.0% (49 of 136), respectively (Fig. 4A). The combination of AFP and our assay resulted in 74.3% sensitivity (101 of 136). In HCC patients with a positive methylated SEPT9 alone (49 patients), BCLC stages 0 and A, the so‐called very early and early stages, accounted for 57.1% (28 of 49) in 4 and 24 patients, respectively, whereas 11, 9, and 1 patients were in HCC stage B, C and D, respectively (Fig. 4B). Therefore, adding the serum CORD assay for methylated SEPT9 enabled us to detect HCC patients, including those with very early and early HCC stages.
Figure 4.
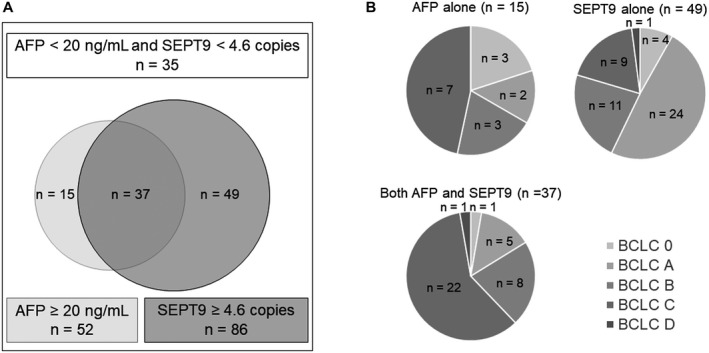
Combination of AFP and serum methylated SEPT9 assays in the HCC group. (A) The positive rate for both assays combined, AFP assay alone, and methylated SEPT9 assay alone was 27.2% (37 of 136), 11.0% (15 of 136), and 36.0% (49 of 136), respectively. The combination of AFP and methylated SEPT9 assays resulted in a sensitivity of 74.3% (101 of 136). (B) HCC stage according to the BCLC staging system in each subgroup (positive based on AFP alone, SEPT9 alone, or both AFP and SEPT9).
Methylated SEPT9 Assay of Tissue Samples
Supporting Table S2 summarizes clinical characteristics for 35 patients with surgically resected HCC. Etiology of HCC was HBV, HCV, and NBNC in 3 (8.6%), 16 (45.7%), and 16 (45.7%) patients, respectively. HCC stage was 0, A, B, and C in 2 (5.7%), 18 (51.4%), 7 (20.0%), and 8 (22.9%) patients, respectively. Histological findings revealed well differentiated HCC, moderately differentiated HCC, poorly differentiated HCC, undifferentiated carcinoma, and combined hepatocellular and cholangiocarcinoma in 6 (17.1%,) 23 (65.7%), 3 (8.6%), 1 (2.9%), and 2 (5.7%) patients, respectively. The median copy number of methylated SEPT9 was 34 (10.85‐51.5) in nontumor tissues (n = 34), and 2,360 (130‐5,860) in tumor tissues (n = 35) (Fig. 5A,B). The median copy number of serum methylated SEPT9 (n = 35) was 6.2 (2.6‐10.0), which was significantly lower than that in tumor tissues (P < 0.001) (Fig. 5C). We subjected 34 paired nontumor and tumor tissue samples to the CORD assay. Nearly all tumor tissues had a significantly higher copy number of methylated SEPT9 than nontumor tissues (P < 0.001) (Supporting Fig. S2).
Figure 5.
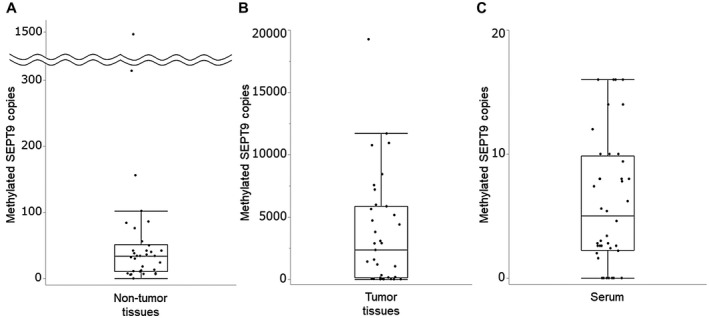
Methylated SEPT9 assay in nontumor tissues, tumor tissues, and serum. (A,B) The median copy number of methylated SEPT9 was 34.0 (10.85‐51.5) in the nontumor tissues (n = 34) and 2,360 (130‐5,860) in the tumor tissues (n = 35). (C) The median copy number of serum methylated SEPT9 (n = 35) was 6.2 (2.6‐10.0), which was significantly lower than that in tumor tissues (P < 0.001).
Discussion
In the present study, we showed that a novel liquid biopsy test targeting methylated SEPT9 is useful for diagnosing HCC. ctDNA is one of the targets in liquid biopsies and carries information on tumor‐specific genetic or epigenetic alterations. DNA methylation is one of these epigenetic alterations, and several studies have investigated ctDNA methylation in various genes.9, 10, 11 In this study, we focused on SEPT9 methylation. SEPT9 acts as either an oncogene or a tumor‐suppressor gene.22 SEPT9 overexpression occurs in human liver tumors23 and rat hepatocarcinogenesis,24 whereas SEPT9 hypermethylation is associated with liver carcinogenesis25 and colorectal carcinogenesis.26 Epi proColon, which targets methylated SEPT9, was the first liquid biopsy test to be approved by the US FDA for colorectal cancer screening12 and has been reported to be useful for diagnosing HCC.13 As an advantage over the Epi proColon assay, the CORD assay requires only a small amount of serum (0.4 mL), because it circumvents bisulfite treatment. Furthermore, the CORD assay is easier to perform, and the methylation level can be evaluated quantitatively (Supporting Table S3). Therefore, we believe the CORD assay is useful for liquid biopsy targeting methylated genes.
The CORD assay uses methylation‐sensitive restriction enzymes and ddPCR. Although there are a few reports on the use of methylation‐sensitive restriction enzymes27, 28, 29 or ddPCR30, 31 for methylation assays, this is the first study to combine both methods for liquid biopsy for HCC. In addition, because a single methylation‐sensitive restriction enzyme may cause incomplete digestion of nonmethylated DNA, which may lead to false PCR amplification, we used three methylation‐sensitive restriction enzymes so that nonmethylated DNA would be completely digested.
The serum methylated SEPT9 CORD assay revealed significantly higher copy numbers in the HCC group than in the healthy control and CLD groups and displayed a sensitivity and specificity of 63.2% and 90.0%, respectively, for diagnosing HCC. Epi proColon has a sensitivity and specificity for HCC of 88.7% and 89.8%, respectively, based on a cutoff value of at least two positive triplicates.13 The proportions of tumor stages differed between our study and this previous Epi proColon report: BCLC stages 0 and A accounted for 45.6% (62 of 136) in our study and for 32.3% (31 of 96) in the previous report. Therefore, we cannot directly compare the clinical performance of our assay with that of Epi proColon 2.0 CE. Furthermore, because Epi proColon is currently not available in Japan, further investigations will be required to compare the clinical performance of the two assays.
We also determined methylated SEPT9 levels in tissue samples. The median copy number was 2,360 in the tumor tissues and 34 in adjacent nontumor tissues. This result was consistent with findings in an earlier report.25 Because the SEPT9 methylation level was significantly different between tumor tissues and adjacent nontumor tissues, we assume that methylated SEPT9 detected in the serum may be derived primarily from tumor tissue. However, because low levels of methylated SEPT9 were found in adjacent nontumor tissues, serum methylated SEPT9 may be partly derived from nontumor hepatic cells associated with CLD.
AFP is the most commonly used surveillance marker for HCC, and the American Association for the Study of Liver Diseases (AASLD) recommends semiannual HCC surveillance using ultrasound imaging with or without AFP assay.3 We also studied the combination of AFP and methylated SEPT9 assays for HCC diagnosis. We used 20 ng/mL as a cutoff value of AFP, as suggested by the AASLD for HCC screening.3 AFP was detected in 39.0% (52 of 136) of patients with HCC, and the assay enabled detecting HCC patients with BCLC stage 0 or A in 8.1% of cases (11 of 136). In contrast, methylated SEPT9 was detected in 63.2% (86 of 136) of patients, and the assay enabled detecting HCC of BCLC stage 0 or A in 25.0% (34 of 136) of patients. In particular, the positive rate of methylated SEPT9 alone was 36.0% (49 of 136), and we could diagnose 28 out of 49 patients based on positive methylated SEPT9 alone (57.1%) as having BCLC stage 0 or A. Therefore, the combination of both assays improved the detection of early HCC over the AFP assay alone. The sensitivity of the AFP assay for HCC diagnosis is low, and our assay can complement this weakness. In addition, there was no significant difference in the positive rate of methylated SEPT9 among patients with NBNC‐HCC and viral‐related HCC (P = 0.672; Supporting Table S1), indicating the same clinical performance of our assay in patients with NBNC‐HCC, including nonalcoholic fatty liver disease–related HCC. Therefore, our assay may be useful for NBNC‐HCC screening for which adequate surveillance systems are currently lacking.3, 4, 5, 6 However, because the sample size was small, further validation in a large population will be required to draw a conclusion.
It has been recently reported that methylated DNA marker panel testing is an effective liquid biopsy to diagnose HCC in an early stage.31, 32 The panel developed by Xu et al.31 had a sensitivity of 83.3% to 85.7% and a specificity of 90.5% to 94.3%, whereas the panel developed by Kisiel et al.32 had a sensitivity of 95% and a specificity of 92%. Both panel tests display high sensitivity and specificity when compared with our assay. However, these panels appear expensive because multiple markers are used, and more than 1 mL of plasma is required. Therefore, we expect that these panels will not be commonly used for HCC screening.
There are several limitations to the present study. First, patients were collected from a few centers, and the cohort size was limited. Therefore, further investigations and a validation study in a large population size are necessary. Second, SEPT9 is not a specific methylation biomarker for HCC,25 and hypermethylation of SEPT9 is associated with various cancers, including colorectal cancer, head and neck squamous cell carcinoma, and breast cancer.22, 33 Thus, an increase in serum methylated SEPT9 may suggest the presence of other types of cancer. We have reported that serum methylated SEPT9 is significantly increased in patients with advanced colorectal adenoma and cancer.17 In the current study, colonoscopy was performed in all of the controls but not in all patients with CLD and with HCC, because it is difficult to perform a colonoscopy in patients who have no symptoms in relation to bowel movement. We suggest that colonoscopy should be indicated for patients with CLD who have elevated methylated SEPT9 levels, to rule out colorectal neoplasms.
In conclusion, the sensitive methylated SEPT9 assay has potential as a liquid biopsy for diagnosing HCC.
Supporting information
Acknowledgment
We thank Ms. Naoko Okayama, Mr. Akihiro Morishige, Mr. Fumiya Takagi, and Mr. Hidekazu Mizuno of the Division of Laboratory, Yamaguchi University Hospital, for their invaluable help in analyzing the data.
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. GLOBOCAN . New Global Cancer Data: GLOBOCAN 2018 | UICC. https://www.uicc.org/news/new-global-cancer-data-globocan-2018. Updated June 7, 2019. Accessed July 1, 2019. [Google Scholar]
- 2. Valery PC, Laversanne M, Clark PJ, Petrick JL, McGlynn KA, Bray F. Projections of primary liver cancer to 2030 in 30 countries worldwide. Hepatology 2018;67:200‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723‐750. [DOI] [PubMed] [Google Scholar]
- 4. European Association for the Study of the Liver . EASL Clinical Practice Guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182‐236. [DOI] [PubMed] [Google Scholar]
- 5. The Japan Society of Hepatology . Clinical Practice Guidelines for Hepatocellular Carcinoma 2013. http://www.jsh.or.jp/English/guidelines_en/Guidelines_for_hepatocellular_carcinoma_2013. Accessed July 1, 2019. [Google Scholar]
- 6. Omata M, Cheng AL, Kokudo N, Kudo M, Lee JM, Jia J, et al. Asia‐Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int 2017;11:317‐370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kudo M. Surveillance, diagnosis, treatment, and outcome of liver cancer in Japan. Liver Cancer 2015;4:39‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van Meer S, de Man RA, Coenraad MJ, Sprengers D, van Nieuwkerk KM, Klümpen HJ, et al. Surveillance for hepatocellular carcinoma is associated with increased survival: results from a large cohort in the Netherlands. J Hepatol 2015;63:1156‐1163. [DOI] [PubMed] [Google Scholar]
- 9. Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, et al. Liquid biopsy in patients with hepatocellular carcinoma: circulating tumor cells and cell‐free nucleic acids. World J Gastroenterol 2017;23:5650‐5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su YH, Kim AK, Jain S. Liquid biopsies for hepatocellular carcinoma. Transl Res 2018;201:84‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer 2019;18:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Potter NT, Hurban P, White MN, Whitlock KD, Lofton‐Day CE, Tetzner R, et al. Validation of a real‐time PCR‐based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem 2014;60:1183‐1191. [DOI] [PubMed] [Google Scholar]
- 13. Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine‐Tresarrieu P, Debard R, et al. Plasma mSEPT9: a novel circulating cell‐free DNA‐based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine 2018;30:138‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raizis AM, Schmitt F, Jost JP. A bisulfite method of 5‐methylcytosine mapping that minimizes template degradation. Anal Biochem 1995;226:161‐166. [DOI] [PubMed] [Google Scholar]
- 15. Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 2001;29:E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suehiro Y, Zhang Y, Hashimoto S, Takami T, Higaki S, Shindo Y, et al. Highly sensitive faecal DNA testing of TWIST1 methylation in combination with faecal immunochemical test for haemoglobin is a promising marker for detection of colorectal neoplasia. Ann Clin Biochem 2018;55:59‐68. [DOI] [PubMed] [Google Scholar]
- 17. Suehiro Y, Hashimoto S, Higaki S, Fujii I, Suzuki C, Hoshida T, et al. Blood free‐circulating DNA testing by highly sensitive methylation assay to diagnose colorectal neoplasias. Oncotarget 2018;9:16974‐16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012;379:1245‐1255. [DOI] [PubMed] [Google Scholar]
- 19. Nygren AO, Dean J, Jensen TJ, Kruse S, Kwong W, van den Boom D, et al. Quantification of fetal DNA by use of methylation‐based DNA discrimination. Clin Chem 2010;56:1627‐1635. [DOI] [PubMed] [Google Scholar]
- 20. Lofton‐Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, et al. DNA methylation biomarkers for blood‐based colorectal cancer screening. Clin Chem 2008;54:414‐423. [DOI] [PubMed] [Google Scholar]
- 21. Wasserkort R, Kalmar A, Valcz G, Spisak S, Krispin M, Toth K, et al. Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer 2013;13:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Connolly D, Abdesselam I, Verdier‐Pinard P, Montagna C. Septin roles in tumorigenesis. Biol Chem 2011;392:725‐738. [DOI] [PubMed] [Google Scholar]
- 23. Scott M, Hyland PL, McGregor G, Hillan KJ, Russell SE, Hall PA. Multimodality expression profiling shows SEPT9 to be overexpressed in a wide range of human tumours. Oncogene 2005;24:4688‐4700. [DOI] [PubMed] [Google Scholar]
- 24. Kakehashi A, Ishii N, Shibata T, Wei M, Okazaki E, Tachibana T, et al. Mitochondrial prohibitins and septin 9 are implicated in the onset of rat hepatocarcinogenesis. Toxicol Sci 2011;119:61‐72. [DOI] [PubMed] [Google Scholar]
- 25. Villanueva A, Portela A, Sayols S, Battiston C, Hoshida Y, Méndez‐González J, et al. DNA methylation‐based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 2015;61:1945‐1956. [DOI] [PubMed] [Google Scholar]
- 26. Song L, Li Y. The role of stem cell DNA methylation in colorectal carcinogenesis. Stem Cell Rev 2016;12:573‐583. [DOI] [PubMed] [Google Scholar]
- 27. Mohamed NA, Swify EM, Amin NF, Soliman MM, Tag‐Eldin LM, Elsherbiny NM. Is serum level of methylated RASSF1A valuable in diagnosing hepatocellular carcinoma in patients with chronic viral hepatitis C? Arab J Gastroenterol 2012;13:111‐115. [DOI] [PubMed] [Google Scholar]
- 28. Hua D, Hu Y, Wu YY, Cheng ZH, Yu J, Du X, et al. Quantitative methylation analysis of multiple genes using methylation‐sensitive restriction enzyme‐based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol 2011;91:455‐460. [DOI] [PubMed] [Google Scholar]
- 29. Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol 2011;91:702‐707. [DOI] [PubMed] [Google Scholar]
- 30. Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Detecting circulating tumor DNA in hepatocellular carcinoma patients using droplet digital PCR is feasible and reflects intratumoral heterogeneity. J Cancer 2016;7:1907‐1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017;16:1155‐1161. [DOI] [PubMed] [Google Scholar]
- 32. Kisiel JB, Dukek BA, Kanipakam RVSR, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, phase I pilot, and phase II clinical validation. Hepatology 2019;69:1180‐1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Matsui S, Kagara N, Mishima C, Naoi Y, Shimoda M, Shimomura A, et al. Methylation of the SEPT9_v2 promoter as a novel marker for the detection of circulating tumor DNA in breast cancer patients. Oncol Rep 2016;36:2225‐2235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


