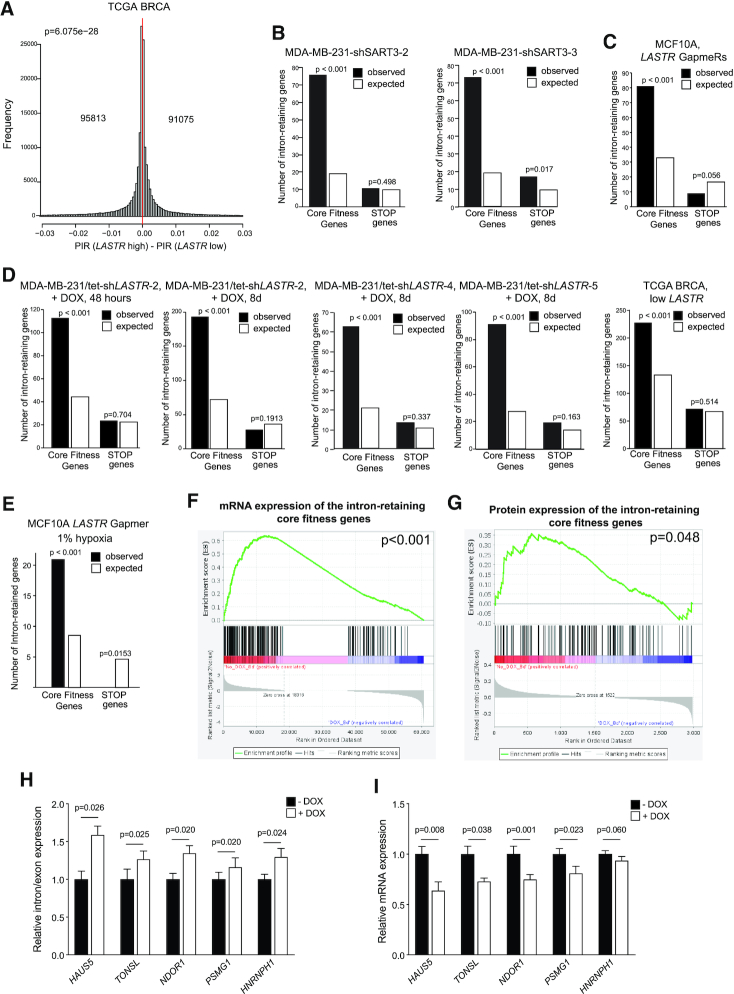
Figure 6.
LASTR loss declines fitness of cancer cells due to decreased expression of essential genes. (A) Histograms showing the distribution of differences in percent of significant intron retention (PIR) in TCGA BRCA tumors with low or high LASTR expression (n = 547). The number of differentially intron retaining genes is shown. P-values were determined by Binomial tests. 25th and 75th percentiles were used to define LASTR high and low expression in the TCGA samples. (B–E) Genes showing increased intron retention in cells with suppressed SART3 or LASTR expression or in the TCGA BRCA samples with low LASTR expression were compared to the lists of core fitness genes (45) or STOP genes (46) using Fisher's exact test. 25th and 75th percentiles were used to define LASTR high and low expression in the TCGA BRCA samples. (F) Gene set enrichment analysis of differentially expressed mRNAs in MDA-MB-231/tet-shLASTR-2 untreated and treated with doxycycline (1 μg/ml, 8 days) among the core fitness genes with significantly increased intron retention. (G) Gene set enrichment analysis of differentially expressed proteins in MDA-MB-231/tet-shLASTR-2 untreated and treated with doxycycline (1 μg/ml, 8 days) among the core fitness genes with significantly increased intron retention. (H, I) RT-qPCR analysis of intron retention/exon expression and mRNA expression levels of the indicated transcripts in MDA-MB-231/tet-shLASTR-2 untreated or treated with doxycycline (1 μg/ml, 8 days). Data are presented as mean ± s.e.m.; P-value was determined by a two-sided t-test, n = 3.