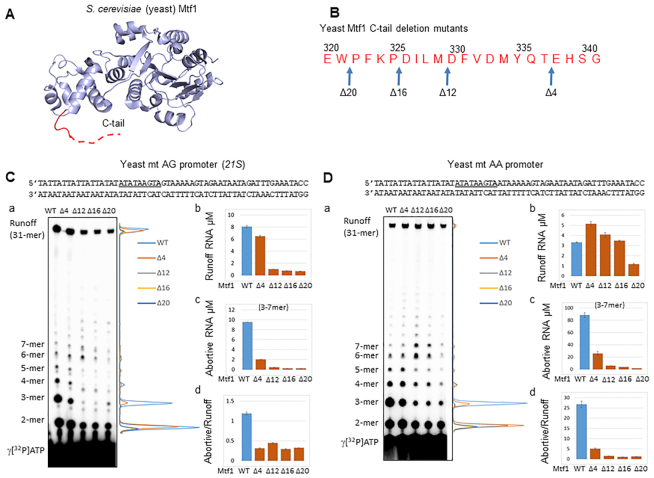
Figure 2.
Deletion of the Mtf1 C-tail reduces abortive RNA synthesis. (A) Structure of the yeast Mtf1 (PDB: 1I4W) with the missing C-tail region in the red dotted line. (B) The amino acid sequence of the yeast Mtf1 C-terminal tail shows the deletion mutants. (C, D) Sequence of the yeast mitochondrial AG and AA promoters (–25 to +32). The nonanucleotide consensus sequence is underlined and start-site is in bold. (a) Image of the polyacrylamide gel (24% polyacrylamide 4 M urea denaturing gel) and the line graphs show the RNA products of the transcription reactions carried out with 1 μM Rpo41, 2 μM Mtf1 and 2 μM promoter duplex for 15 min using 250 μM ATP, UTP, GTP and 1.25 mM 3′dCTP spiked with γ[32P]ATP. Panels b, c, and d show the μM amounts of runoff RNA, 3–7 mer abortive RNA, and abortive to runoff ratio on the AG promoter and AA promoter. Error bars are from two measurements.