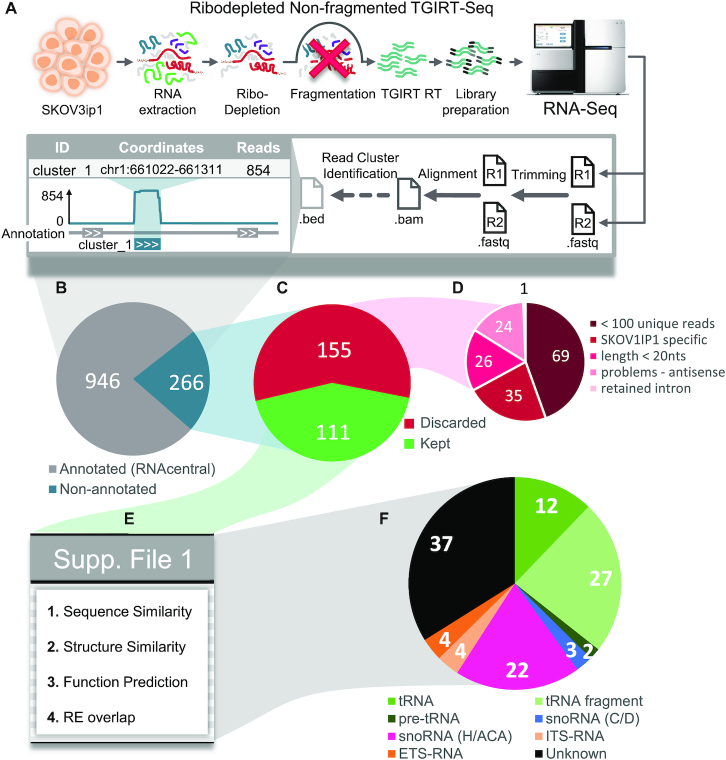
Figure 1.
Identification of new non-coding RNA genes from sequencing read clusters. (A) Description of the RNA-Seq and bioinformatic analysis pipelines used for RNA detection and identification. The lack of fragmentation is indicated by an X. R1 and R2 indicate forward and reverse read files obtained from paired-end sequencing. Read clusters were identified using Blockbuster on bam alignment files, and visualized on a genome browser. (B) Identification of new non-coding RNA clusters. The RNA clusters obtained from A were compared to the annotation available in RNAcentral and clusters with and without standard annotations are indicated in the pie chart. (C) Proportion of kept and discarded clusters. Clusters with <100 uniquely mapped reads, a size smaller than 20 nucleotides, not detected in other investigated TGIRT-Seq datasets, poorly defined antisense fragment of the 45S rRNA or representing a retained intron were discarded. It should be noted that clusters with fewer than 100 uniquely mapped reads but with multimapped reads only aligning to other non-annotated clusters were kept (such is the case for the ETS and ITS clusters). (D) Distribution of the discarded clusters non-optimal features. (E) Methods used for the functional (biotype) classification of the new non-coding RNA genes. The retained clusters were compared to the sequence, structure, function and repeated element overlap of known ncRNA and classified by biotype. (F) Final distribution of the predicted biotype of the NA_RNA clusters.