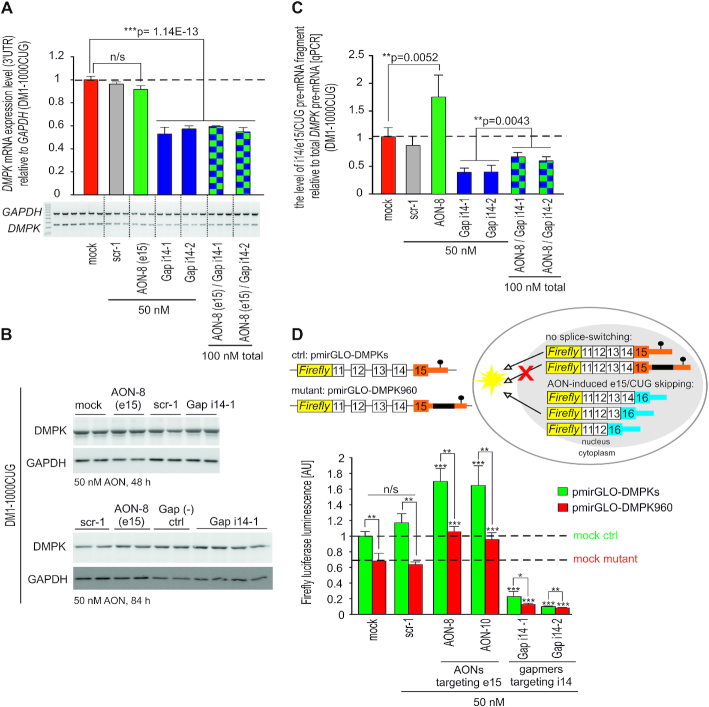
Figure 3.
AON-mediated skipping of e15 triggers accumulation of spliced out pre-mRNA fragment containing CUG-expansions and mitigates nuclear retention of DMPK. (A) RT-PCR quantitation of total DMPK mRNA in DM1-1000CUG fibroblasts 72 h post transfection with indicated AONs (PCR amplicon within 3′UTR). Mock, scrambled sequence AON (scr-1) and gapmer AONs were used as controls and representative RT-PCR image is shown below bar graph. (B) Representative immunoblots showing no changes in DMPK protein expression in DM1 fibroblasts 48 h (upper gel) and 84 h (bottom gel) post AON dosing. (C) qPCR quantitation of i14/e15/CUG spliced-out pre-mRNA fragment vs total DMPK pre-mRNA in DM1-1000CUG fibroblasts 72 h after dosing the cells with e15 splice-switching AON-8 either alone or in a combination with i14-1 and i14-2 gapmer AONs targeting distinct regions of i14. Scrambled-sequence AON scr-1 and gapmer AONs were used as controls. i14/e15/CUG pre-mRNA was amplified with primers located within i14 and e15 (upstream of CUG-repeat region), primers for DMPK pre-mRNA amplification were located within i9 and e10. (D) Outline and results of the firefly luciferase reporter assay modeling the DM1 translational defect. Luciferase reporter constructs (schematic) with a 3′UTR harboring genomic DMPK fragment spanning e11–e15 along with entire 3′UTR carrying either expanded (pmirGLO-DT960) or no repeats (pmirGLO-DMPKs) were transfected into Cos7 cells and dosed with indicated AONs. Mock and scr-1 were used as controls. Horizontal dashed lines indicate luciferase expression level in control and mutant mock treated samples.