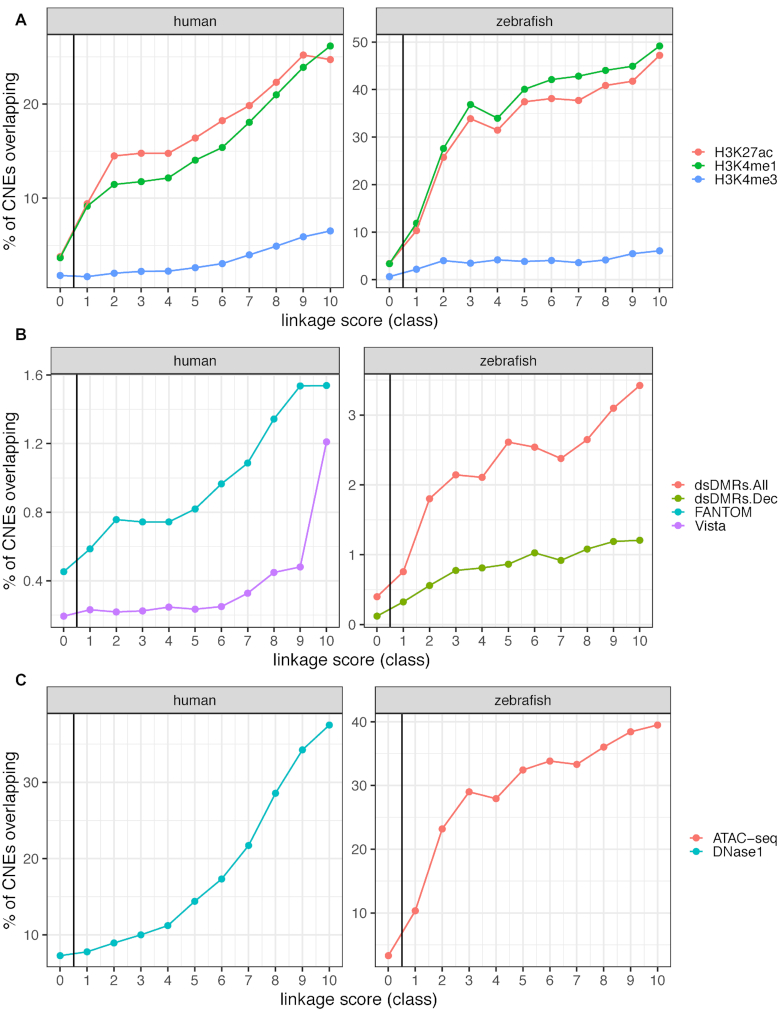
Figure 2.
Overlap between PEGASUS CNEs and functional annotations. (A) Percentage of CNEs overlapping histone modification ChIP-seq peaks from embryonic stem cells in human (left) (10) and various developmental stages in zebrafish (right) (36). (B) Percentage of CNEs overlapping enhancer predictions from FANTOM5 (24) or Vista (37) in human (left) and from differentially methylated regions during development in zebrafish (right). ds.DMR stands for developmental stage-specific differentially methylated regions, regions which exhibit tissue-specific differences in methylation levels and overlap distal cis-regulatory regions (38). dsDMRs.All: all sites at all stages; dsDMRs.Dec: sites with decrease in methylation from 6 to 24 hpf embryos, shown to be specific to early development (38). (C) Percentage of CNEs overlapping open chromatin regions, DNase1 peaks in human embryonic stem cells (10) (left) and overlap of ATAC-seq regions with a normalized read count higher than five in zebrafish embryos (39) (right). CNEs were divided into 10 deciles of equal size according to their linkage scores. Class 0 represents CNEs with no associated target gene.