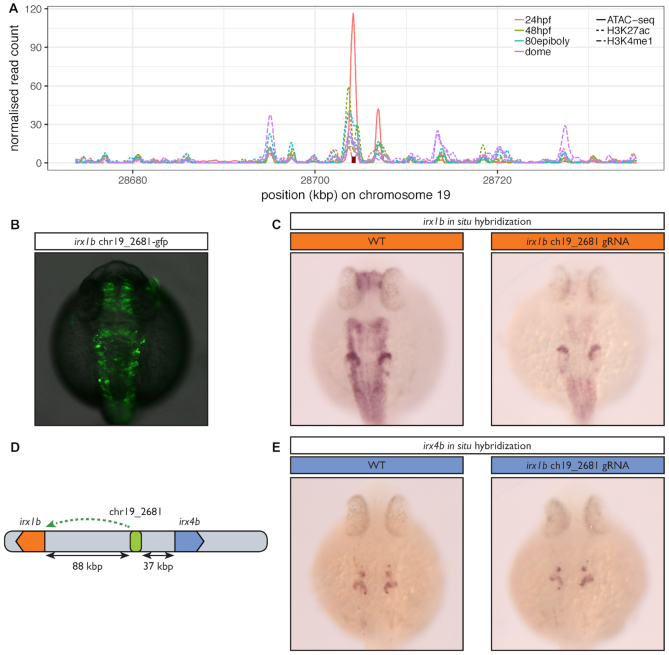
Figure 6.
In vivo inactivation of a predicted ancestral enhancer for irx1b affects its expression. (A) Evidence for the regulatory potential of the chr19_2681 CNE. The figure shows the normalized read counts for a ChIP-seq analysis of histone modifications (H3K4me1 & H3K27ac) in four developmental stages (dashed & dotted lines) (36) and for an ATAC-seq analysis in 24 hpf embryos (full lines) (39) in a 60 kb region around chr19_2681 (red rectangle). (B) 24 h old F0 zebrafish embryos injected with a Tol2 transposon containing the predicted irx1b CNE positioned 5′ of the gata2 minimal promoter driving green fluorescent protein (GFP) expression. (C) In situ hybridization for irx1b mRNA performed on 24 h old wild type embryos (WT) or embryos injected with a mix of three CRISPR/Cas9 ribonucleoprotein complexes targeted at the predicted irx1b enhancer. The CNE activity profile overlaps with the expression profile of irx1b, which comprises the acousticovestibular ganglia, the caudal diencephalon, the tectum, the hindbrain, the spinal cord and the anterior part of the otic vesicle but not the mid-hindbrain boundary. irx1b expression level is greatly decreased in all these structures when the CRISPR/Cas9 complex is targeted to the CNE compared to the control, establishing it as a bona fideirx1b enhancer. (D) The chr19_2681 CNE, predicted to target irx1b is located closer to irx4b (37 kb) than to irx1b (88 kb). (E) In contrast to (B), irx4b’s expression profile which includes the anterior part of the otic vesicle and a few cells in the hindbrain is not affected by the CRISPR/Cas9 complex showing that this CNE is specific to irx1b and does not regulate irx4b.