Figure 1.
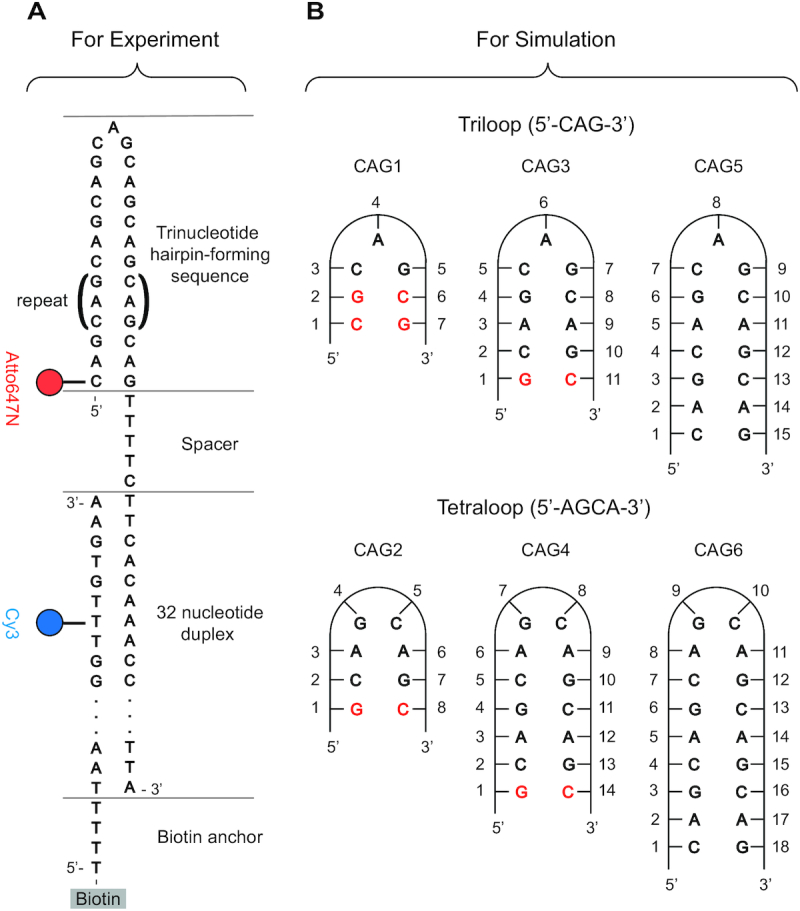
(A) Schematic DNA design for the smFRET analysis of CAG repeat hairpins, with donor in blue and acceptor in red. The CAG sequence in parenthesis is repeated for various hairpins. The hairpin loop of interest is immobilized to a surface by a partial complimentary DNA anchor strand. The TTTTC spacer helps reduce the interaction between the hairpin and junction duplex. (B) CAG hairpins sequences considered in the MD simulations. CG Watson-Crick pairs were added to the ends of short repeats to mimic long canonical stems.
