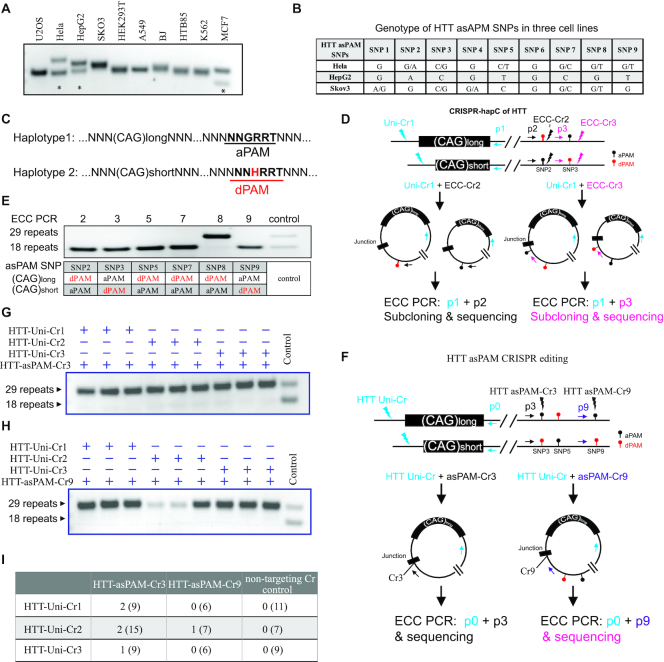
Figure 4.
Haplotype-specific HTT editing achieved with CRISPR-hapC and asPAM CRISPRs. (A) Genotyping of CAG expansion in exon 1 of the HTT gene in 10 cell lines. (B) Genotyping of the nine HTT asPAM SNPs in three cell lines by sequencing. (C) Schematic illustration of two haplotypes comprising the HTT CAG expansion allele and the asPAM allele. Here, the active PAM (aPAM) is linked to the long CAG allele. (D) Schematic illustration of HTT haplotyping by the CRISPR-hapC approach. Uni-Cr1, a CRISPR that cleaves both alleles. ECC-Cr2 and ECC-Cr3, CRISPR target sites adjacent to the asPAM SNP and used for generating extrachromosomal circular (ECC) DNA. (E) ECC-PCR-based haplotyping of the HTT CAG expansion allele with HTT asPAM SNP2, 3, 4, 7, 8 and 9. (F) Illustration of HTT asPAM CRISPR editing and genotyping with the eccDNA method. The p0, p3 and p9 indicate genotyping primers of ECC-DNA encompassing the ECC-DNA junction. (G) and (H) ECC-DNA PCR genotyping for haplotype-specific deletion of the long CAG expansion allele (29 repeats). Transfections were conducted with a pair of CRISPRs: one universal CRISPR that cleave both alleles and one asPAM-specific CRISPR that only cleave the active PAM allele linked to the long CAG expansion (n = 3). Controls are CRISPR pairs that delete both haplotypes. (I) Summary of single cell colonies edited with a pair of HTT-Uni-Cr and HTT-asPAM-Cr as shown in the table. Note that genotyping was conducted with PCR primers flanking the targeted HTT deletion region. All edited clones have the long CAG allele deleted. Numbers in parentheses are the total number of clones screened for each group.