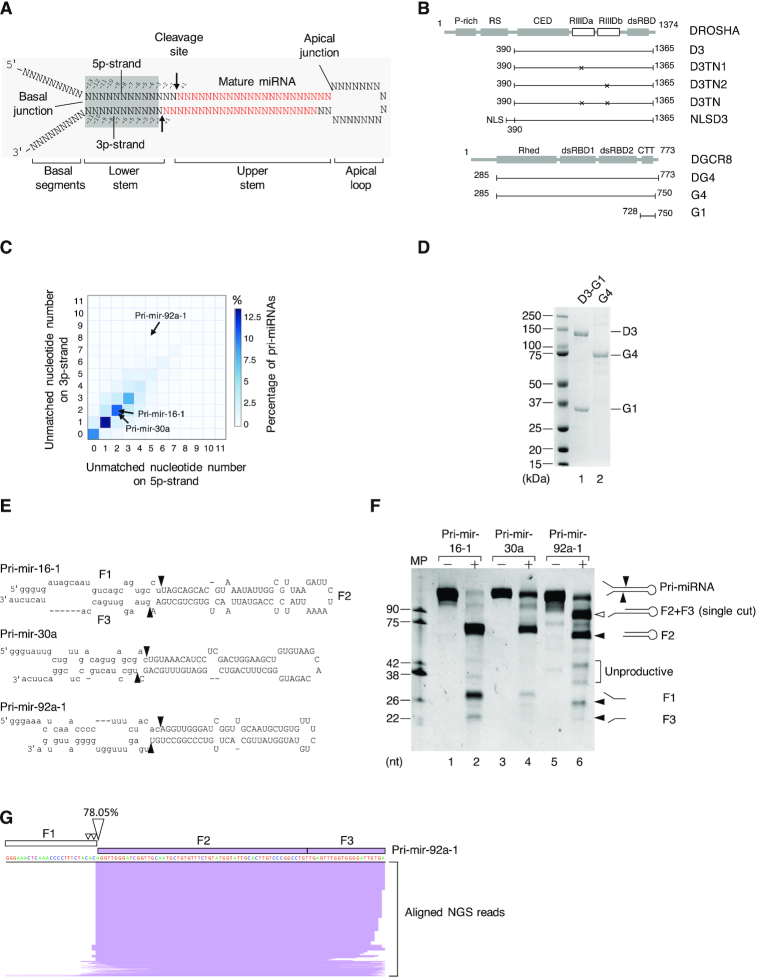
Figure 1.
The Microprocessor complex executes a single cleavage on the 5p-strand of pri-miRNAs. (A) Schematic illustration of the pri-miRNA structure. The mature miRNA region is shown in red. The arrows indicate the cleavage sites of Microprocessor. (B) The protein domain structure of DROSHA and DGCR8. P-rich: Proline-rich domain; RS: Arginine/serine-rich domain; CED: central domain; RIIIDa and RIIIDb: RNase III (a and b) domains; dsRBD: double-stranded RNA-binding domain; Rhed: RNA-binding heme domain; CTT: C-terminal tail region; and NLS: Nuclear localization sequence. (C) The percentage of human pri-miRNAs containing different numbers of unmatched nt in their lower stems. The unmatched nt for the 5p- and 3p-strands of the pri-miRNAs were quantified as described in the Materials and Methods. (D) The purified D3-G1 complex and the G4 dimer analyzed by SDS-PAGE. (E) Schematic illustration of pri-mir-16-1, pri-mir-30a, and pri-mir-92a-1. (F) The pri-miRNA processing assays. Five pmol of each pri-miRNA were incubated with 8 pmol each of D3-G1 and the G4 dimer for 2 h at 37°C. (G) The long product (F2+F3) in (F) was cloned and sequenced using next-generation sequencing (NGS), after which NGS reads were aligned with the pri-mir-92a-1 sequence and visualized by IGV. MP: Microprocessor.