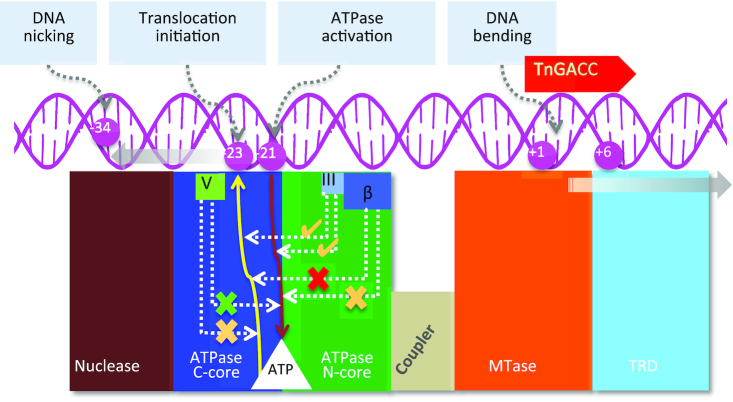
Figure 7.
Schematic representation of DNA-mediated coupling of the ATPase to translocase and nuclease activities. The target sequence (+1 to +6) and the region of DNA bending upon target binding, which steers the DNA towards the ATPase domain, are shown. The continuous brown arrow from the DNA at -21 to the ATP binding pocket (white) indicates the length of DNA required for full stimulation of the ATPase, while the yellow line to the DNA at –23 indicates DNA length required for initiation of ATPase-coupled DNA translocation. The effect of the deletion of the β-hairpin loop, and mutation of motif III LlaBIII-Thr376 and motif V LlaBIII-Arg564 on the ATPase and translocase activities are illustrated using dotted arrow lines connecting the respective structural elements to the brown and yellow arrows illustrating DNA dependent ATPase and translocase activities. A tick mark indicates detectable nucleolytic cleavage, while a cross mark indicates no cleavage. An amber tick mark indicates a diminished ATPase or translocase but detectable nuclease activities, the green and the amber cross in the case of motif V indicates an active ATPase and a diminished translocase activities resulting in an inactive nuclease. The red and amber cross in the case of β-hairpin deletion mutant indicates reduced ATPase activity but inactive translocase and nuclease activities. The solid grey arrows indicate the direction of translocation of the DNA and the Type ISP RM enzyme, with respect to each other. -34 is the position of nicking by a static enzyme (20).