Table 1.
Kinetic and thermodynamic parameters for Zur–DNA interaction in E. coli cells (error bars are s.d.)
| ZurmE |
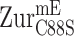
|
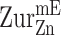
|
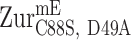
|
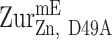
|
|
|---|---|---|---|---|---|
| k 1(nM−1 s−1)a | 0.80 ± 0.07 | 0.77 ± 0.08 | 0.46 ± 0.08 | 0.68 ± 0.24 | 0.55 ± 0.08 |
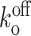 (s−1) (s−1) |
25 ± 12 | 22 ± 21 | 5.4 ± 0.6 | 22 ± 2 | 36 ± 41 |
| k r (s−1) | 16 ± 11 | 12 ± 20 | n/ob | 21 ± 1 | 27 ± 40 |
| k f (nM−1 s−1) | 0.012 ± 0.002 | 0.018 ± 0.003 | 0.011 ± 0.014 | 0.021 ± 0.006 | 0.026 ± 0.004 |
| K m (nM) | 14 ± 10 | 12 ± 17 | n/ob | 39 ± 18 | 7.6 ± 4.5 |
K
d1 ( =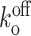 /k1) (nM)a /k1) (nM)a
|
31 ± 15 | 28 ± 27 | 12 ± 3 | 33 ± 12 | 67 ± 48 |
| K d2 ( = k-2/k2) (nM)a | 990 ± 80 | 830 ± 200 | 1300 ± 400 | 500 ± 160 | 1300 ± 300 |
| K d3 ( = k-3/k3)a | 0.011 ± 0.002 | 0.023 ± 0.007 | 0.022 ± 0.023 | 0.032 ± 0.062 | 0.008 ± 0.006 |
| [D0]NB (nM)a | 2700 ± 200 | 2300 ± 500 | 2900 ± 700 | 2000 ± 500 | 3700 ± 800 |
| [D0]TB⋅no (nM)a | 100 ± 2 | 82 ± 8 | 130 ± 30 | 75 ± 12 | 92 ± 9 |
a n o = 5 was used in fitting; see Supplementary Figure S15 for no dependence of parameters.
[D0]NB: concentration of nonspecific binding sites in cell. [D0]TB: concentration of tight binding sites in cell.
bNot observed.
