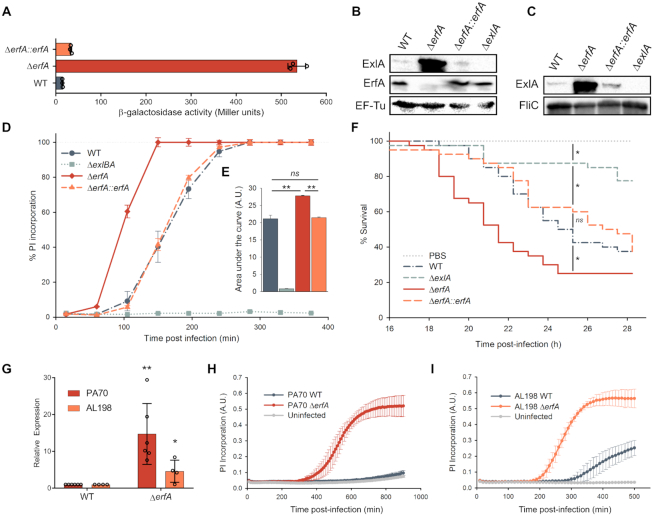
Figure 2.
ErfA regulates ExlA-dependent virulence through inhibition of exlBA expression in P. aeruginosa. (A) β-Galactosidase activities of IHMA87 strains harboring exlBA::lacZ. The strains were grown in LB, and activities of the transcriptional fusion were measured at OD600 = 1. The enzyme activities are represented as mean ± standard deviation (SD) from three independent experiments. (B andC) Immunodetection of ExlA in bacterial cell extracts (B) and supernatants (C). Whole bacteria and culture supernatants were sampled and analyzed by SDS-PAGE for ExlA, ErfA, EF-Tu and FliC content using appropriate antibodies. (D) Kinetics of bacterial cytotoxicity on epithelial cells. A549 epithelial cells grown in 96-well plates were infected with indicated strains at a multiplicity of infection (MOI) of 10 in the presence of propidium iodide (PI). PI incorporation, which reflects membrane permeabilization, was monitored every 45 min by automated live-cell microscopy and normalized to the total number of cells measured at the start of the experiment by Syto24 staining. Data are represented as mean ± s.e.m. from three independent experiments. (E) The areas under each curve presented in (D) were calculated from the values obtained after 380 min of infection. ns: not significant. (F) Survival curves of Galleria mellonella larvae infected with an average of 5 bacteria per larva. Forty larvae were infected per strain. Significance testing was performed using log-rank test (P < 0.05). (G) RT-qPCR analysis of exlA expression in PA70 and AL-198 strains and erfA isogenic mutants. All strains were grown in LB medium to OD600 = 1. Expressions were normalized to the abundance of rpoD mRNA. Error bars indicate the SD. The P-value was determined using one-tailed T-test and is indicated by * (P < 0.05) or ** (P < 0.01). (H andI) Kinetics of PI incorporation (A.U.: Arbitrary Unit) into A549 epithelial cells infected at a MOI of 10 with PA70 (H) and AL-198 (I) strains and isogenic erfA mutants.