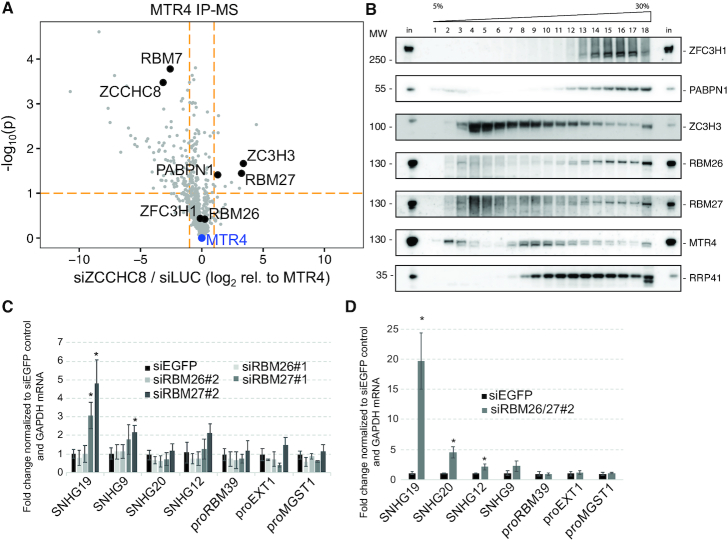
Figure 3.
RBM26 and RBM27 harbor PAXT activity. (A) Comparison of protein abundance (label-free quantification (LFQ) values) in triplicate hMTR4-LAP affinity capture mass spectrometry (AC-MS) experiments from ZCCHC8-depleted (siZCCHC8) to control (siLUC) HeLa Kyoto cells. Volcano plot depicts the ratio of protein abundances (log2(LFQ)) normalized to the MTR4 bait from siZCCHC8 versus siLUC AC-MS experiments (X-axis) and the -log10 of the P-values from a Student’s t test for this comparison. Only proteins significantly enriched relative to a control AC-MS in either siLUC or siZCCHC8 (see Supplementary Figure S3A and S3B) are depicted here. Relevant proteins are highlighted and labeled. Orange dashed lines mark log2FC = -/+ 1 and -log10P-values = 1. (B) Glycerol gradient (5–30%) western blotting analysis of the sedimentation of the indicated proteins from extracts of HEK293 ZC3H3-FLAG protein expressing cells. Fractions of input (‘in’) were loaded on first and last lanes for comparison. (C) RT-qPCR analyses of PAXT and NEXT substrates as in Figure 2B but using total RNA harvested from RBM26-depleted (siRBM26), RBM27-depleted (siRBM27) or control (siEGFP) HeLa cells. Results of two independent RBM26 and RBM27 siRNAs (indicated as #1 and #2) are shown. Data are displayed as mean values, with error bars denoting SD (n = 4 biological replicates). ∗ P < 0.05, Student’s t test. (D) RT-qPCR analyses of PAXT and NEXT substrates as in panel (C) using total RNA harvested from RBM26/RBM27 (siRBM26/27) co-depleted or control (siEGFP) HeLa cells. Note that for co-depletion only siRNAs #2 were used. Data are displayed as mean values, with error bars denoting SD (n = 3 biological replicates). ∗ P < 0.05, Student’s t test.