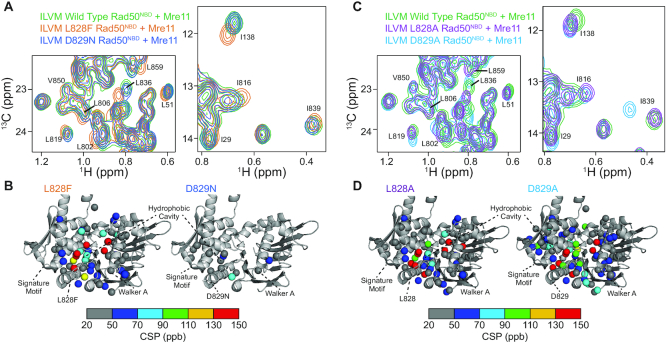
Figure 4.
D-loop mutations disparately affect Rad50 structure. (A) Regions of the overlaid 2D 13C,1H HMQC spectra for U-2H12C, Ileδ1–13CH3, Leuδ/Valγ-[12CD3/13CH3], Metϵ-13CH3 (ILVM)-labeled wild type (green), L828F (orange) and D829N (blue) Rad50NBD bound to unlabeled Mre11 (see Supplementary Figure S5 for the full spectra). The labeled peaks experience a chemical shift perturbation (CSP) indicating a change in structure upon D-loop mutation. (B) Methyl group resonances that have a significant CSP are mapped on the structure of Rad50 (pdb: 3QKS (15)) as spheres. CSPs in the L828F and D829N mutants are shown on the left and right structures, respectively, and site of each mutation is depicted with sticks. Methyl groups with small (<20 ppb) or no CSP are hidden. The color of the sphere indicates the relative structural change, as defined by the gradient below the structures. (C) Regions of the overlaid 2D 13C,1H HMQC spectra for ILVM-labeled wild type (green), L828A (purple), and D829A (light blue) Rad50NBD bound to unlabeled Mre11 (see Supplementary Figure S5 for the full spectra). The labeled peaks experience a chemical shift perturbation (CSP) indicating a change in structure upon D-loop mutation. (D) Methyl group resonances that have a significant CSP are mapped on the structure of Rad50 (pdb: 3QKS (15)) as spheres. CSPs in the L828A and D829A mutants are shown on the left and right structures, respectively, and site of each mutation is depicted with sticks. Methyl groups with small (<20 ppb) or no CSP are hidden. The color of the sphere indicates the relative structural change, as defined by the gradient below the structures.