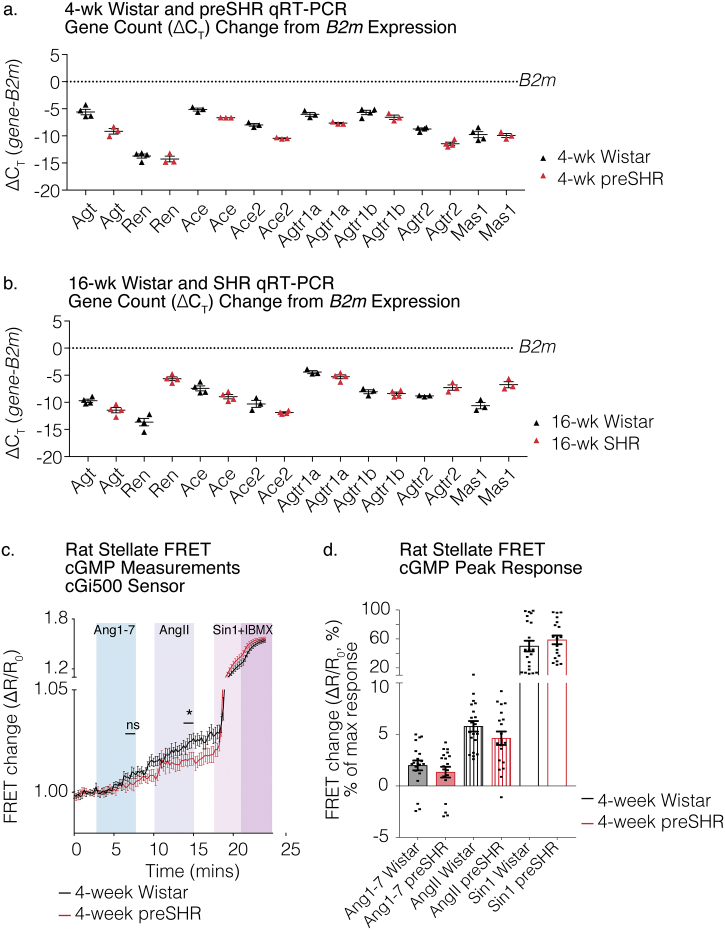
Fig. 4.
Angiotensinergic mRNA transcript validation by qRT-PCR in rat stellate ganglia.
The presence of the RNA transcripts Agt, Ren, Ace, Ace2, Agtr1a, Agtr1b, Agtr2 and Mas1 was confirmed by qRT-PCR in sympathetic stellate ganglia from four-week Wistar and preSHR ganglia (a), and 16-week adult Wistar and SHR (b). The qRT-PCR raw counts were first normalized to a control gene B2m as per the comparative (∆CT) method [48]. Each data point corresponds to one stellate RNA sample from one rat. Data are displayed as ∆CT mean ± SEM. FRET microscopy was conducted on sympathetic stellate neurons obtained from Wistar (n = 11 rats, 3 cultures, 20 cells) and preSHR rats (n = 9 rats, 3 cultures, 19 cells). Cells were transduced with the cGi500 FRET sensor and randomly selected for imaging. Increases in cGMP generation was observed in sympathetic neurons in response to Ang1–7 and AngII (c, d). Maximal FRET changes were evoked following administration of a combination of the NO-donor Sin-1 (10 μM) and the PDE inhibitor IBMX (100 μM). There was significantly greater cGMP generation in response to AngII in Wistar vs. preSHR neurons (two-way ANOVA, p = .0403). Peak FRET changes were obtained in response to AngII or Ang1–7 and converted to percentage FRET changes and values are depicted as a proportion of the maximal FRET change (%). There was no difference in peak FRET responses in response to Ang1–7 or between strains (d). Data are displayed as mean ± SEM.