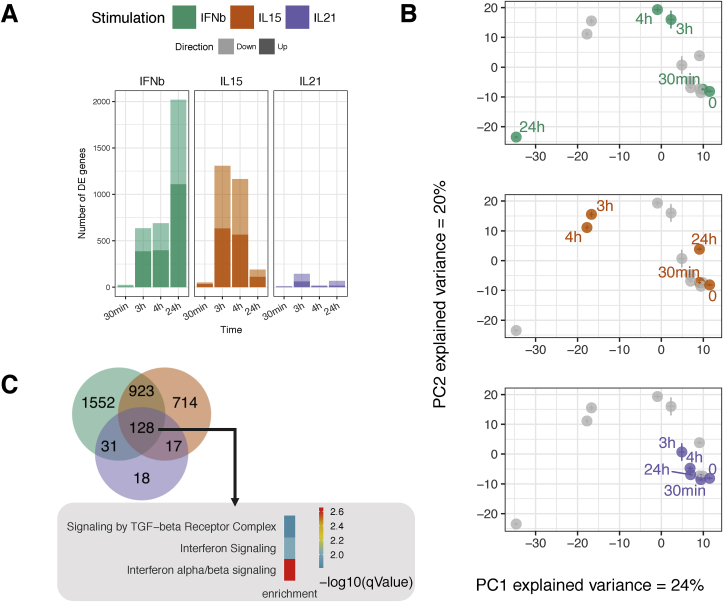
Fig. 1.
Distinct and common transcriptional response of IE-CTLs in response to tissue alarmins and adaptive cytokine. After 12 days in culture, IE-CTLs were starved for 48 h, then stimulated for 30 min, 3 h, 4 h or 24 h with IFNβ, IL-15 or IL-21. Unstimulated cells (0 h) were used as control. n = 8 samples per time point and condition. The transcriptome was analyzed by RNA-seq. (A) Number of DEGs after cytokine treatment in comparison with the unstimulated samples at each time point (|log2 fold change|>1, FDR ≤ 0.01). Light colors indicate up-regulated genes. Dark colors indicate down-regulated genes. (B) Centroids of all the samples using Principal Components 1 and 2 from all DEGs reveal different time course patterns of gene expression upon IFNβ (green), IL-15 (orange) or IL-21 (purple) stimulation on IE-CTLs. (C) Venn diagram depicting the number of unique and overlapping genes across the stimulations and time points. The most significantly enriched pathways (determined by Reactome) of shared genes between the three stimulations are depicted. Color (key) indicates level of significance. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)