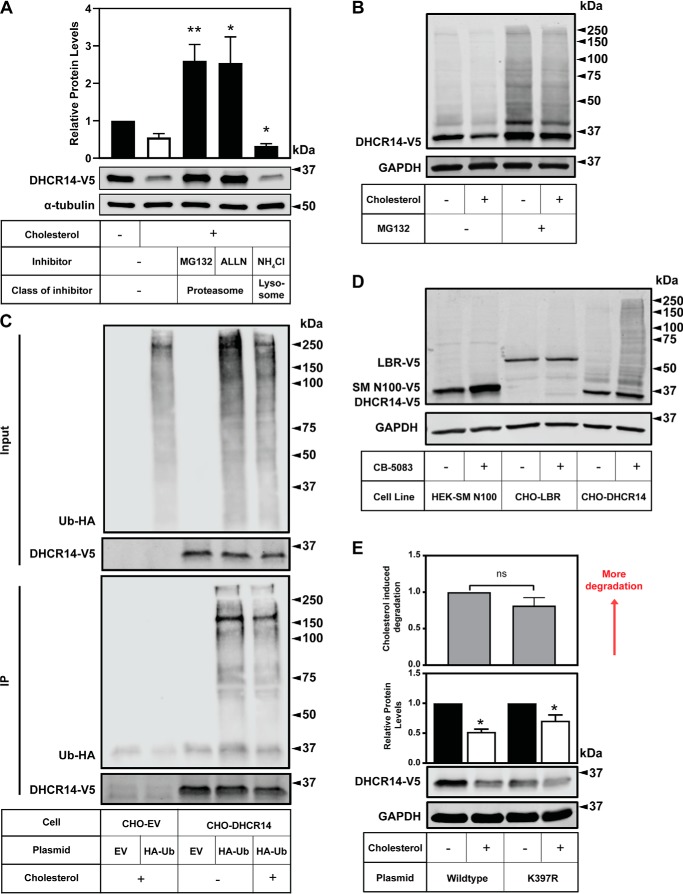
Figure 6.
DHCR14 is degraded by the proteasome and is ubiquitinated, although the published ubiquitination site is not responsible for DHCR14 sterol-mediated turnover. A, CHO-DHCR14 cells were treated for 8 h with or without 20 μg/ml Chol/CD, with either MG132 (10 μm), ALLN (25 μg/ml), or NH4Cl (20 mm). B, CHO-DHCR14 cells were treated for 8 h with or without 10 μm MG132 and with or without 20 μg/ml Chol/CD, and cell lysates were harvested. C, CHO-EV and CHO-DHCR14 cells were transfected with empty vector or HA-tagged ubiquitin plasmids for 24 h. Cells were treated for 2 h with 10 μm MG132 to block proteasomal degradation and subsequently treated with 10 μm MG132 with or without 20 μg/ml Chol/CD for 4 h. Cell lysate was immunoprecipitated with V5 antibody conjugated to magnetic dynabeads. D, HEK293-SM N100-GFP-V5, CHO-LBR, and CHO-DHCR14 cells were treated for 8 h with or without the VCP inhibitor CB-5083. E, CMV-DHCR14-V5 or CMV-DHCR14-K397R-V5 was transiently transfected into CHO-7 cells for 24 h. Transfected cells were treated for 8 h with or without 20 μg/ml cholesterol complexed to cyclodextrin. Cell lysates or IP products were separated by SDS-PAGE, and protein levels were quantified by Western blotting using V5 (DHCR14), HA (HA-ubiquitin), and α-tubulin or GAPDH. Data are presented as mean ± S.E. (error bars) of n = 5 (A) or n = 6 (E) independent experiments or presented as representative blots of n = 4 (B), n = 2 (C), and n = 1 (D) for previously published SM-N100 (40), n = 3 for LBR-V5, and n = 6 for DHCR14-V5. A paired Student's two-tailed t test was used for statistical analysis. *, p < 0.05; **, p < 0.01; ns, nonsignificant.