Figure 8.
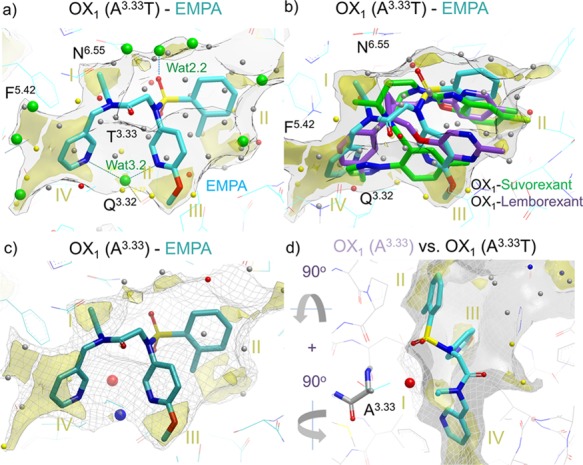
Role of water in EMPA (8) OX1/OX2 selectivity. (a) EMPA ligand in OX2 mimicking OX1 A1273.33T mutant with a WaterFLAP-computed network (small spheres, color-coded by energy) and X-ray crystallographic waters (large green spheres). The interstitial water hydrogen bonding to the two pyridine nitrogens and Q1263.32 can be clearly seen. (b) Suvorexant (2) and lemborexant (4) ligand poses from OX1 crystal structures overlaid with an EMPA water network in OX2. The carbon atoms of the ligands are colored cyan (EMPA), green (suvorexant), and purple (lemborexant). GRID maps are contoured (transparent solid) and colored in the following manner: C1 is the probe (lipophilic) in yellow at −2.8 kcal/mol, and the CH3 methyl group probe is in gray at 1 kcal/mol, which defines the pocket surface in terms of how close a ligand carbon atom can reside. WaterFLAP water networks calculated on the pseudo-apo structure (shown as large spheres) have been color-coded in red if predicted to have a free energy (ΔG) >3.5 kcal/mol, in yellow if ΔG is between 2.0 and 3.5 kcal/mol, in gray if ΔG is between −1.0 and 2.0 kcal/mol, and in blue if ΔG < −1.0 kcal/mol. All WaterFLAP free energy estimations are relative to bulk solvent. (c, d) Comparison of the binding site surfaces of the OX2 mimicking the OX1 A1273.33T mutant structure (solid) and back mutated T1273.33A/wild-type (WT) OX1 (dark gray mesh) indicates that in wild-type OX1, an energetically unhappy water molecule will be trapped by the OX2-selective EMPA antagonist. WaterMap water network calculations of the complex with OX1 (A1273.33) with a very unhappy (high relative energy to bulk solvent, 4 kcal/mol) water trapped in the larger OX1 binding site, shown as a large red sphere. The water stabilized by the two pyridines is also shown as a large blue sphere (stabilized, 2 kcal/mol); in the pseudo-apo structure, this water is calculated by WaterFLAP to be unstable relative to bulk water (small yellow sphere in panel (b)).
