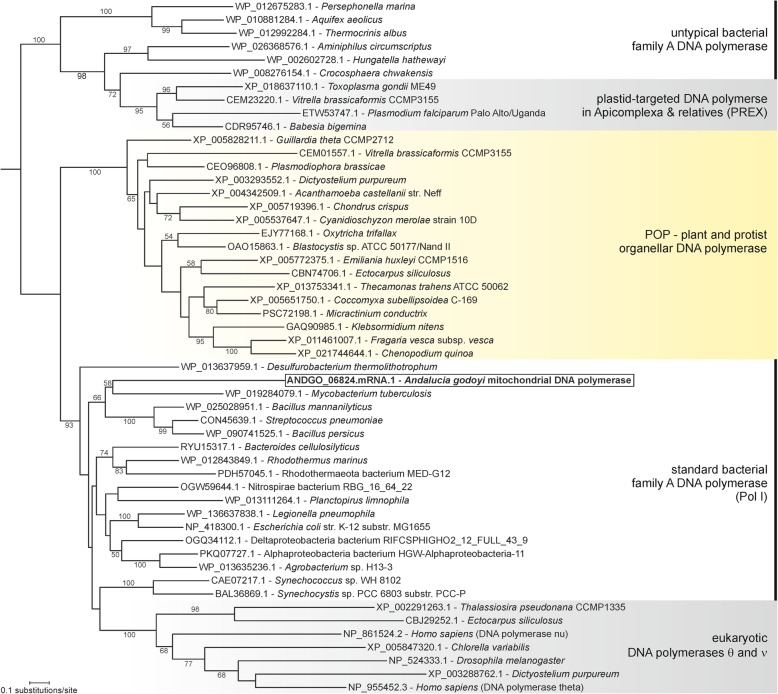
Fig. 3.
Phylogenetic analysis of family A DNA polymerases. The arbitrarily rooted ML tree was inferred using RAxML (PROTGAMMALG model) based on an alignment of 533 amino acid positions. Branch support values (rapid bootstraps) ≥ 51% are shown. Note the position of the mitochondrion-targeted A. godoyi DNA polymerase, branching off among standard bacterial family A DNA polymerases (Pol I) and separately from the organellar (mitochondrial or dually targeted) DNA polymerases widespread in plants and protists (the POP group)