Extended Data Figure 2 |. CRL1β-TRCP RBX1 RING and CUL1 WHB domains, without or with a covalently-linked NEDD8, are dynamic in the absence of other factors and are harnessed in catalytic architecture for substrate ubiquitylation with UBE2D.
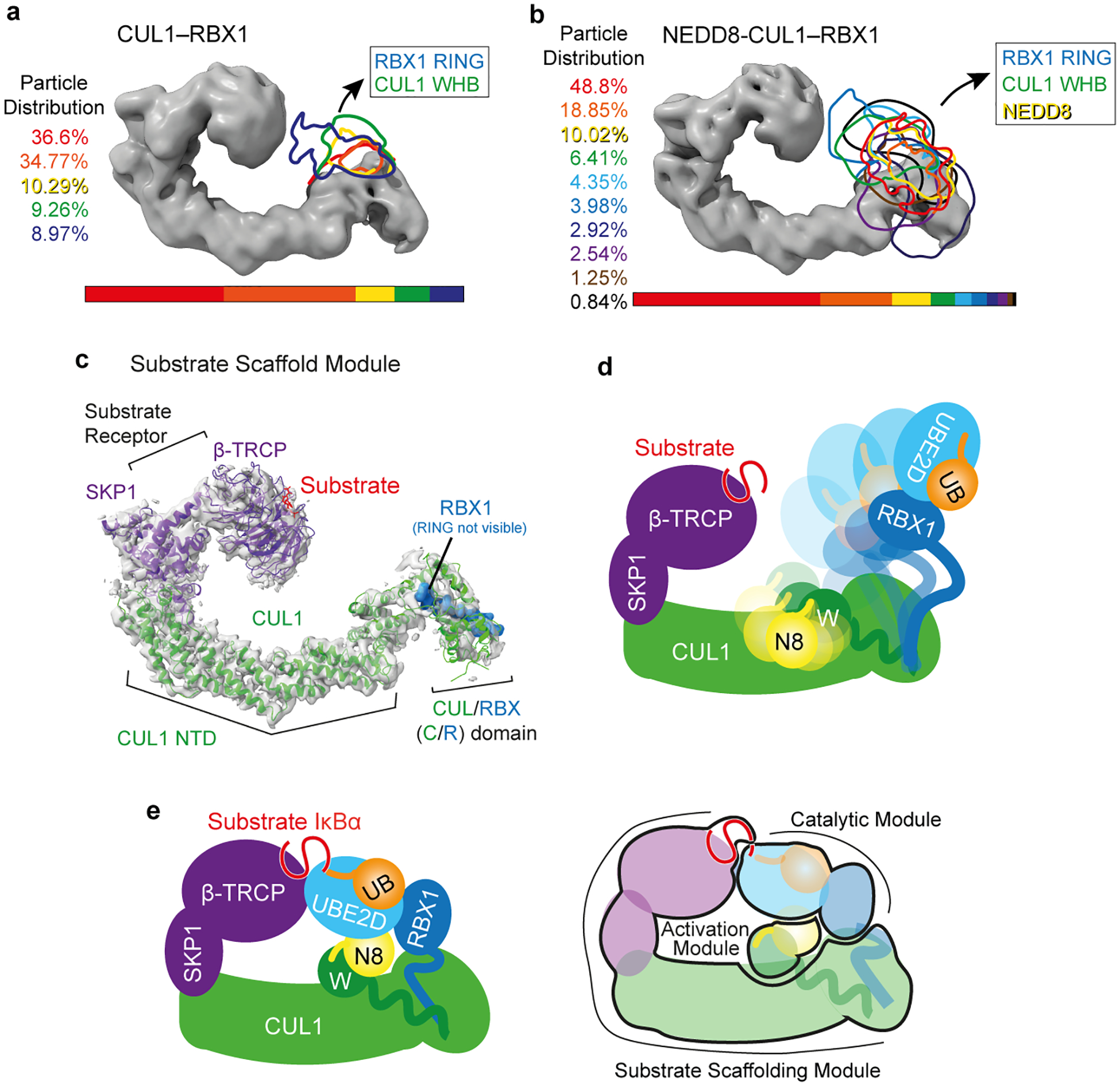
a, Cryo EM density corresponding to substrate scaffolding regions of unneddylated CRL1β-TRCP is shown as surface with that encompassing RBX1’s RING and CUL1’s WHB domains, outlined for different 3D classes in different colors corresponding to percent of particles in that 3D class. b, same as in a, but with neddylated CRL1β-TRCP, with its surfaces outlined in different classes encompassing RBX1’s RING, CUL1’s WHB domain, and covalently modified NEDD8. c, Refined cryo EM density from CRL1β-TRCP reveals substrate scaffolding module bridging the substrate recruited to substrate receptor β-TRCP with the intermolecular cullin-RBX (C/R) domain, readily fitted with crystal structures of SKP1-β-TRCP [PDB: 1P22]20 and CUL1’s N-terminal domain and the C/R domain of CUL1-RBX1 [PDB: 1LDK]7. d, Cartoon showing dynamics of NEDD8, its linked CUL1 WHB domain, and the RBX1 RING domain based on cryo EM data in b for substrate-bound neddylated CRL1β-TRCP, and model for varying locations of the RBX1 RING-bound UBE2D~UB relative to the substrate awaiting ubiquitylation. e, Left, cartoon representation of the catalytic architecture based on the cryo EM data shown in Figure 2, representing neddylated CRL1β-TRCP-catalyzed UB transfer from E2 UBE2D to an IκBα-derived substrate peptide. Right, semi-transparent version of the cartoon, highlighting the three modules (substrate scaffolding, catalytic, and activation modules), their constituents and locations establishing the catalytic architecture for substrate priming by neddylated CRL1β-TRCP and UBE2D.
