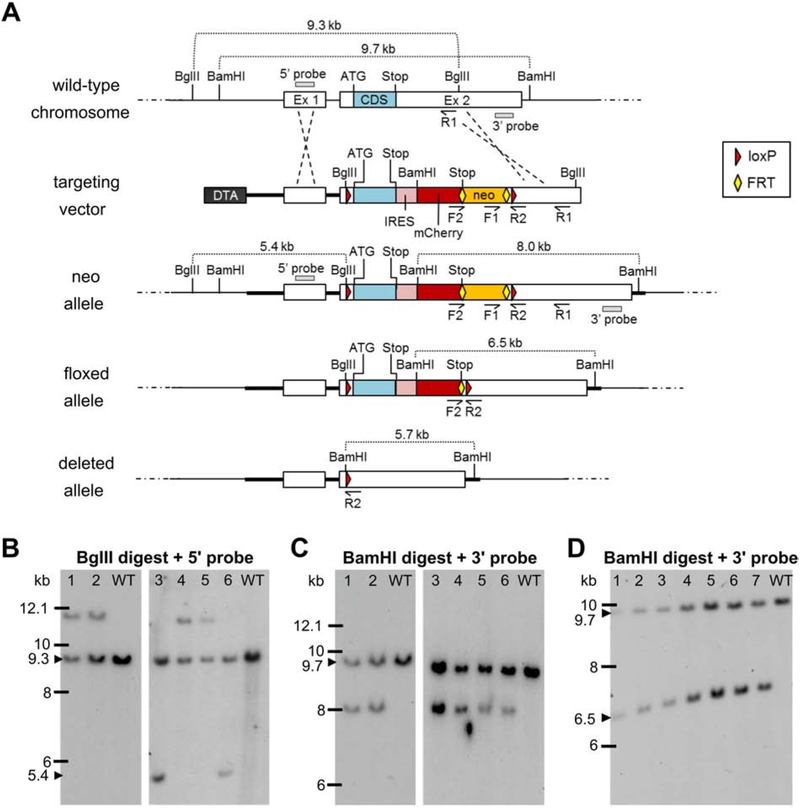
Figure 1.
Targeting strategy for generation and verification of the floxed Kcna1 allele. A, Utilizing subcloned genomic sequences from the endogenous wild-type Kcna1 chromosomal region, a targeting vector was generated containing two loxP sites (red triangles) flanking the exon 2 (Ex 2) coding sequence (CDS); a Diphtheria Toxin A (DTA) negative selection marker; a neomycin (neo) cassette positive-selection marker flanked by two FRT sites (yellow diamonds); and an internal ribosomal entry site (IRES)-mCherry fluorescent reporter. The targeting construct was introduced into embryonic stem (ES) cells to obtain the neo allele by homologous recombination. Recombined ES cell clones were then used to generate chimeric males which were mated to females with Flp recombinase to excise the neo cassette in the germline, eventually yielding mice with the floxed allele following another round of breeding. In the presence of Cre recombinase, the CDS and IRES-mCherry reporter of the floxed allele are removed to yield the deleted allele. The F1/R1 primer pair was used to screen ES cells by PCR for 3’ homologous recombination. The F2/R2 primer pair was used to identify by PCR heterozygous floxed mice with Flp-mediated excision of the neo cassette. Restriction enzyme sites for BgIII and BamHI are indicated, as well as the locations of the 5’ and 3’ probes used for Southern blotting. B-C, Southern blots of ES cell DNA digested with BgIII and BamHI to test for 5’ and 3’ homologous recombination, respectively, to yield the neo allele. Clone numbers 3 and 6 exhibited restriction fragments indicative of successful 5’ and 3’ recombination events. D, Southern blot of BamHI-digested genomic DNA from seven heterozygous floxed mice verifying Flp-mediated excision of the neo cassette.