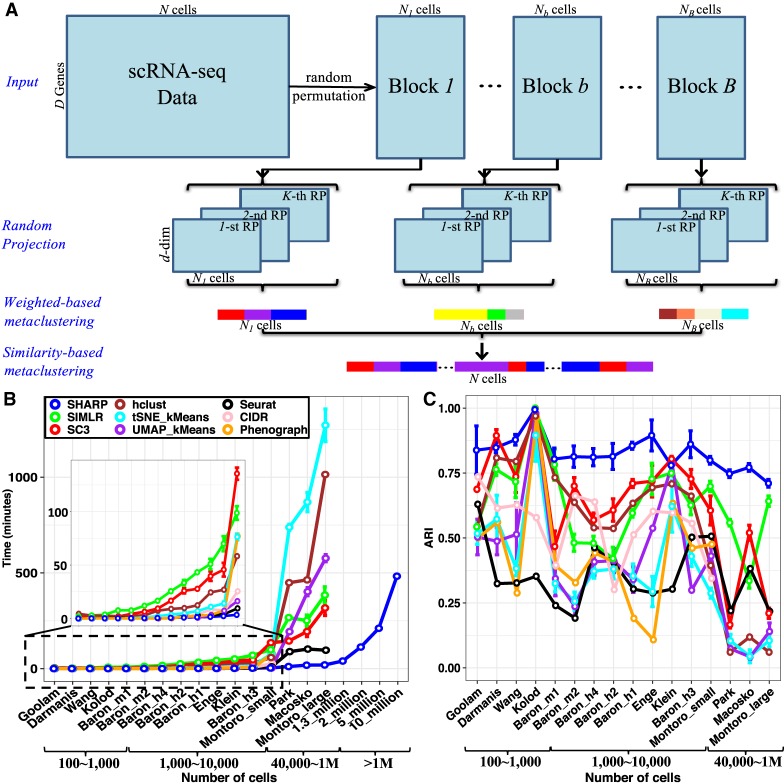
Figure 1.
The framework of SHARP. (A) SHARP has four steps for clustering: divide-and-conquer, random projection (RP), weighted-based metaclustering, and similarity-based metaclustering. (B,C) Running time (B) and clustering performance (C) based on ARI (Hubert and Arabie 1985) of SHARP in 20 single-cell RNA-seq data sets with numbers of single cells ranging from 124 to 10 million (where data sets with 2 million, 5 million, and 10 million cells were generated by randomly oversampling the data set with 1.3 million single cells). For the data sets with more than 1 million cells, only SHARP can run, and only the running time was provided owing to lack of the ground-truth clustering labels. All of the results for SHARP were based on 100 runs of SHARP on each data set. All the tests except for the larger-than-1-million-cell data sets were performed using a single core on an Intel Xeon CPU E5-2699 v4 @ 2.20-GHz system with 500-GB memory. To run data sets with more than 1 million cells, we used 16 cores on the same system. CIDR and PhenoGraph were unable to produce clustering results for those data sets with number of cells larger than 40,000 (i.e., Park, Macosko, and Montoro_large).